Jonathan Kane recently (Oct. 6, 2017) posted an article on The Panda's Thumb where he claimed that Young Earth Creationists often don't get enough credit for raising serious issues about evolution [Five principles for arguing against creationism].
He mentioned some articles about pseudogenes as prime examples. I asked him for references and he responded with two articles by Jeffrey Tomkins that were published on the Answers in Genesis website. The first was on the β-globin pseudogene and the second was on the GULO pseudogene. Both articles claim that these DNA sequences aren't really pseudogenes because they have functions.
I'll deal with the β-globin pseudogene in this post and the GULO pseudogene in a subsequent post.Here's the article ....
Before addressing the specific criticisms in this article it's important to not lose sight of the bigger issue. Creationists tend to focus on particular examples while ignoring the big picture. In this case, there is abundant evidence of gene duplications in all species and there's abundant evidence that the fate of one duplicated copy of a gene is often to become inactivated rendering it a pseudogene. This has given rise to a robust explanation of multigene families referred to as Birth-and-Death Evolution [The Evolution of Gene Families] [On the evolution of duplicated genes: subfunctionalization vs neofunctionalization]. In order for Young Earth Creationists to mount a serious challenge to evolution they need to provide a better explanation for all this data and they need to provide solid evidence that the Earth is less than 10,000 years old.The Human Beta-Globin Pseudogene is Non-Variable and Functional.
Jeffrey P. Tomkins, Institute for Creation research, Dallas, TX, USA
Answers Research Journal
Abstract: One of the iconic (yet enigmatic) arguments for human-ape common ancestry has been the β-globin pseudogene (HBBP1). Evolutionists originally speculated that apparent mutations in HBBP1 were shared mutational mistakes derived from a human-chimpanzee common ancestor. However, others noted that if the gene was indeed non-functional, then it should have mutated markedly in the past 3 to 6 million years of human evolution due to a lack of selective constraint on the region. Recent research confirms that the HBBP1 region of the 6-gene β-globulin cluster is highly non-variable compared to the other β-globin genes based on large-scale DNA diversity assessment within both humans and chimpanzees. Highlighting the lack of HBBP1 sequence variability is genetic data from three different reports that link point mutations in the HBBP1 gene with β-thalassemia disease pathologies. Biochemical evidence for functionality is indicated by multiple categories of functional genomics data showing that the HBBP1 gene is transcriptionally active and a key interactive component of the β-globin gene network. In brief, the HBBP1 gene encodes two consensus regulatory RNAs that are alternatively transcribed and/or post-transcriptionally spliced. This functional complexity produces at least 16 different exon variant transcripts and 42 different intron variant transcripts. Two major regulatory regions in the HBBP1 locus contain active transcription factor binding sites that overlap multiple categorical regions of epigenetic data for functionally active chromatin. The HBBP1 gene also has the most regulatory associations with active and open chromatin within the entire β-globin cluster and its transcripts are expressed in at least 251 different human cell and/or tissue types. Instead of being a useless genomic fossil according to evolutionary predictions, the HBBP1 gene appears to be a highly functional and cleverly integrated feature of the human genome that is intolerant of mutation.
There are about 15,000 pseudogenes of various kinds in the human genome. You can't challenge the big picture of pseudogenes and junk DNA by picking out one example and trying to prove it has a function. This will not refute evolution even if it turns out to be true that one particular stretch of DNA looks like a pseudogene but actually has a function. And it certainly won't be evidence of a Young Earth.
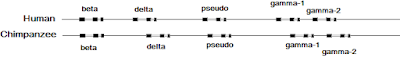
Now let's deal with the Tomkins article. Here's a diagram showing the pseudogene in the β-globin gene cluster in humans and chimps.
There's a pseudogene at this locus in most of the great apes—an observation that's consistent with a duplication event tens of million of years ago followed by the loss of function of one of the copies. The pseudogene became fixed in the ancestral population and was passed down to all modern species. The rate at which most of the pseudogene sequence has accumulated base substitutions is consistent with the rate at which neutral mutations are fixed by random genetic drift. This indicates that most of the sequence is not under negative selection. As far as I know, creationists—especially Young Earth Creationists—haven't offered a reasonable explanation of this observation.
Tomkins' main point is that this stretch of DNA has a function so, presumably, the creator(s) copied this useful part of the genome and plugged it into one of the chromosomes as they were building each of the species. They didn't really care very much about the surrounding DNA so they didn't worry about copying it exactly. As it turns out, they introduced differences in the surrounding DNA so that the sequences of chimps and humans differ by about 2% and chimps and gorillas differ by about 4%. Humans and gorillas also differ by about 4%. The important point is that there are far fewer differences in the exons of the functional genes so they look "conserved" if you adopt an evolutionary perspective.
There's a stretch of DNA near the human β-globin pseudogene that has far fewer changes if you examine the chimp and human genomes. In evolutionary terms, it is "conserved." (It is reusable design if you are a Young Earth Creationist.) Tomkins quotes a paper by Moleirinho et al. (2013) documenting this conservation. The explanation is that the region between the γ-globin genes and the pseudogene is involved in regulating expression of the β-globin genes, probably because it contains a scaffold attachment site and associated sequences that regulate chromatin conformation. This role appears to have arisen shortly before the divergence of chimps and humans.
Here's what the sequence similarities look like on the UCSC Genome Browser. The degree of sequence similarity between the human genome and the genomes of chimps, gorillas, orangutans, and monkeys is shown as a histogram where the height of the bar indicates significant similarity. As you can see, the exons of the functional genes are conserved but the pseudogene sequence is not conserved. This is exactly what you expect if the pseudogene sequence is gradually drifting away from the ancestral gene that was functional right after the gene duplication event.
Much of the sequence surrounding the γ-globin genes is under selection, including a stretch that extend toward the pseudogene. This is the regulatory region that controls expression of the entire locus.
Thus, the evolutionary explanation is that a gene duplication occurred and one of the copies became a pseudogene. Subsequently, a region in the vicinity of the pseudogene acquired a new function involved in chromatin looping and regulation. That's why a large stretch of DNA near the γ-globin gene is conserved. I don't know how Tomkins explains the data other than just saying that the presence of function casts doubt on evolution.
Tomkins' other evidence for function relies on the ENCODE data. He notes that the pseudgogene region is transcribed as part of the pervasive transcription noted by ENCODE. It also contain numerous transcription factor binding sites, DNase I sensitive regions, and histone markers. Some of this might be remnants of the original gene but most are just spurious events that occur throughout the genome in junk DNA. Sandwalk readers will be familiar with the idea that ENCODE data does not prove function.
UPDATE:
John Harshman send me his comparison of the β-globin region on the UCSC Genome Browser.
As you can see, the pseudogene region seems to be only slightly less conserved than the functional genes in this analysis. This isn't unexpected. The functional genes will drift apart over 100 million years by accumulating neutral mutations in the coding regions. The pseudogene arose about 65 million years ago in primate ancestors so it will have accumulated mutations at a faster rate since that time but not before. The difference in the primate lineage should amount to about 20% in that time.
When you compare the "conservation" of the various loci using an outgroup to the primate lineage, the pseudogene will only be about 20% less conserved than the functional genes. That's pretty much what you see in the figure.
When you do a binary comparison (e.g. chimp vs human), I'm assuming the algorithm subtracts the neutral mutation rate in order to calculate whether a sequence is conserved or not. Thus, in my figure, the pseudogene region only shows up as a small blip. This may be statistical error or a small bit of conserved sequence within the the second exon.
That's how I interpret the results. Any help will be appreciated. If you know how to get % sequence similarity comparisons on this browser then please post that information in the comments or email me.
Moleirinho, A., Seixas, S., Lopes, A.M., Bento, C., Prata, M.J., and Amorim, A. (2013) Evolutionary constraints in the β-globin cluster: the signature of purifying selection at the δ-globin (HBD) locus and its role in developmental gene regulation. Genome Biology and Evolution 5:559-571. [doi: 10.1093/gbe/evt029]



67 comments :
I'm a bit confused about the last figure. You write that the degree of similarity between the genomes is shown as the variation in height of the bar. But how can one bar be thicker than the others?
It isn't obvious to me how, for example, the Rhesus monkey bar can have a short area where it is thicker than the others. Is it then more similar to the four others than they are to each other?
To me it looks as if Moleirinho et al. were documenting conservation as well as low within-population diversity in the pseudogene itself, not near it, though the pattern is odd. The introns apparently are more conserved than the exons, and neither is as conserved as the typical functional protein-coding sequence. It's hard to reconcile the UCSC track with the numbers in the paper. Could you explain further?
Tomkins is so very confused.
He starts by writing "One of the iconic (yet enigmatic) arguments for human-ape common ancestry has been the β-globin pseudogene (HBBP1). Evolutionists originally speculated that apparent mutations in HBBP1 were shared mutational mistakes derived from a human-chimpanzee common ancestor."
The implication here is that evolutionists predicted that the gene was a pseudogene because of a lack of conservation. So Tomkins basically affirms that the prediction from evolution is based on sequence conservation.
Then he writes: "Recent research confirms that the HBBP1 region of the 6-gene β-globulin cluster is highly non-variable compared to the other β-globin genes based on large-scale DNA diversity assessment within both humans and chimpanzees."
So he's saying the locus is actually "conserved", without really giving any measures on how much change counts as conservation. But if that is actually true, that it is conserved over evolutionary time, then the prediction from evolution isn't that it's a junk gene.
Also, am I correct in my reading that he seems to be talking about two different measures of conservation? The between-species level of conservation, and the within-population level. That's two very different things and they are definitely not equivalent. Tomkins looks to be trying to argue that the evolutionary prediction based on between species conservation has been falsified because there is a much smaller among population level variability for the locus. Am I reading this right?
Assuming he's doing it "correctly" and is talking about between-species divergence: Obviously you work with the kind of data you have. If you have data that indicates that a region is not conserved between species, but divergent at a rate consistent with neutral drift, then that would lead you to predict it's probably junk. But if you then later learn that your previous data was wrong (which must be what Tomkins is claiming?), and the gene is not consistent with a neutrally evolving region, then the prediction would obviously change on that basis. After all, the prediction is based on the degree of conservation.
So when Tomkins writes "Instead of being a useless genomic fossil according to evolutionary predictions, the HBBP1 gene appears to be a highly functional and cleverly integrated feature of the human genome that is intolerant of mutation." - he's actually just setting up a strawman by pretending that the evolutionary prediction was based on something else than the conservation data.
Now, I get from your post that Tomkins is really just wrong, the gene isn't actually conserved but seems to be evolving at a rate consisten with neutral accumulation of mutations.
The paper is quite confusing but it does seem like the authors are claiming less diversity within the pseudogene itself. I haven't found anything in the literature to confirm their results but there are papers that discuss the putative regulatory sequences between the pseudogene and the gamma-globin genes.
Moleirnho et al. conclude that the region around the pseudogene is probably involved in chromatin and protein interactions required for regulation.
The important point is that the pseudogene is still a pseudogene. It resembles a beta-globin gene but it doesn't make any globin. There's no solid evidence that the pseudgene sequences themselves have acquired a secondary function.
There's no solid evidence that the pseudgene sequences themselves have acquired a secondary function.
Moleirinho et al. claim to present such evidence, in both sequence conservation and reduced nucleotide diversity. How do you refute that?
Thanks for posting this article, but you've misrepresented what I was arguing at Panda's Thumb. I never claimed that creationists should get credit for raising serious issues about evolution. I'm confident that the vast majority of their objections to evolution are baseless. My point has always been that if we don't point out in detail why their objections are baseless, and instead dismiss their arguments as not worthy of a response, in the long run it's going to result in more examples of the sort of outcome described in Sharp and Bergman's book Persuaded by the Evidence. I really thought I was clear about what I was arguing in my article there, so it bothers me that my viewpoint has been so grossly misunderstood.
I may respond to the rest of this post at a later point, but for now the most important thing is to clear up this misunderstanding. Do you understand now what my perspective actually is?
The Panda's Thumb?! I thought that site was dead.
The similarity is to the human genome (which is not shown). Apparently at that aligned location the Rhesus monkey shows more similarity to the human genome than do the other primates.
I'm not REFUTING the data in the Moleirinho et al paper. I'm just not accepting it as solid evidence for function within the pseudogene until the work is confirmed and a function identified.
I'm still trying to figure out the apparent contradiction between Moleirinho et al. and the UCSC track. Can you advise?
Are you Jonathan Kane? 'Cause if you are I don't think I've misrepresented your position. In fact, I agree with it.
You argued repeatedly in the comments that some creationist papers have not been adequately addressed by the anti-creationists. You are quite critical of evolution supporters for not taking the time to refute some of those articles. The only logical conclusion is that some of those creationists deserve a bit more respect because their arguments are not trivial.
I agree. That's why I've spent a considerable amount of effort over the years trying to address issues raised by the likes of Jonathan Wells, Michael Behe, Michael Denton, and Stephen Meyer ... to name just a few. Like you, I'm often disappointed in the responses from the evolution side because they often misunderstand and misrepresent the arguments from these creationists.
They may be wrong but they deserve more credit for coming up with strong arguments to defend their position—arguments that the majority of evolution defenders can't refute because they lack sufficient knowledge.
It stumbles and falls from time to time but it always get back up.
Pandas at the Toronto Zoo
Yes, I'm Jonathan Kane. Sorry if that wasn't clear; I always use my Google account when I comment at Blogspot.
I suppose it's your wording that I was objecting to. The statement that I think creationists are raising "serious issues about evolution" suggests that I think their arguments are valid. For the most part they aren't, but that doesn't mean their arguments should be ignored. I'm glad you agree with this.
By the way, of the five papers that I linked to in my response to you at Panda's Thumb, I hope someone will eventually address the one arguing that the first portions of the chimpanzee genome to be sequenced were contaminated with human DNA. That's the only one of them that hasn't received any kind of response yet.
When I look at the UCSC genome browser and set up a track comparing sequence conservation in human, chimp, gorilla, and orangutan, I find a conserved area between HBD and HBG1, right where the pseudogene ought to be. It's about as conserved as either of its neighbors. I don't see why that doesn't appear on your track.
I don’t know why either.
Can you make a link that will show the comparison on our screens too?
Could it be that that region is conserved in human/chimp/gorilla, but not if you look at a broader set of species including other apes and rhesus macacques?
Oddly enough, according to the UCSC Genome Browser histogram Larry posted, the whole beta-globin gene cluster locus in Green Monkeys, which should be the most distantly related to Homo Sapiens, is also the one that is on average the most similar to the Homo S. locus. The bar is thicker across it's entire length compared to the other ones.
I added gibbon, rhesus, and green monkey to the track. No significant change.
"Evolutionists originally speculated that apparent mutations in HBBP1 were shared mutational mistakes derived from a human-chimpanzee common ancestor."
I'd like to know why he refers to "evolutionists" ... ah! propaganda tactics, sorry.
"However, others noted that if the gene was indeed non-functional, ..."
Wait a second. Shared disabling mutations are right there. That means that, if translated, the proteins produced would be dysfunctional, not that the gene, or the regions, itself has no functions at all. This is a misdirection. The issue, that there's shared mutations disabling the potential protein was not even touched. Why keep reading? Ah! For fun. OK.
"... In brief, the HBBP1 gene encodes two consensus regulatory RNAs ..."
It doesn't encode anything. These regulatory RNAs are not translated.
" ... Two major regulatory regions in the HBBP1 locus contain active transcription factor binding sites that overlap multiple categorical regions of epigenetic data for functionally active chromatin."
Still, nothing about the inactivated potential protein.
"... Instead of being a useless genomic fossil according to evolutionary predictions, the HBBP1 gene appears to be a highly functional and cleverly integrated feature of the human genome that is intolerant of mutation."
I don't see anything clever about a disabled protein coding region that is used, instead, as regulatory RNA and binding region for transcription factors. The disabled potential protein is still there. Mutations that disable it are still shared between humans and chimps. So distracting ourselves with RNA and regulatory regions doesn't explain why those disabling mutations are shared between humans and chimps, other than because we inherited the mutations from a common ancestor.
Is the GULO thing like this? A distraction from the issue?
There's a recent paper in Genes & Development showing that deleting the HBBP1 gene in a human hematopoietic cell line results in increased expression of gamma globin. That seems like evidence of function to me.
Also re Moleirinho. I don't think the authors are saying that the pseudogene is more conserved overall than HBB but that the pseudogene is more conserved overall than the intronic sequence of HBB. Also the conservation within the pseudogene is pretty equally distributed between exonic and intronic sequence while the conservation in HBB is much higher in exons than in introns (as you'd expect for a protein coding gene).
None of which of course supports creationism in the slightest.
Funny thing: Tomkins claims that he couldn't find a sequence alignment for the region under discussion, but then he references the UCSC genome browser, where such an alignment is easily available. Cargo cult science.
"Shared disabling mutations are right there. That means that, if translated, the proteins produced would be dysfunctional, not that the gene, or the regions, itself has no functions at all. This is a misdirection. The issue, that there's shared mutations disabling the potential protein was not even touched."
I don't think that works. If the sequence is non-functional then the disabling mutations not only are strong evidence for common ancestry but indicate natural mutations destroying a gene product. But if the pseudogene has function then IDers can always claim the pattern of disabling mutations indicates 'common design'
Nothing will work with them either way.
I think it is very important to understand what's been "claimed" by "evilutionists," namely, that those mutations render the potential protein useless. The region still looks like a region coding for a globin. This thing about the region having a function is a mere distraction. The "claim" has never been "this is a pseudogene with no functions whatsoever." The regions still contains a pseudogene with respect to the protein coding portions. It still looks convincingly like a broken beta-globing coding gene. The breaking still shares commonalities between humans and chimps. Case closed. This regulatory RNA and binding is mere distraction from the actual point.
Interestingly, Jonathan Kane was saying that creationists update their stuff. False. This is the very same "argument" against vestigial organs, but at a different level. There's a function! There's a function! Maybe. Let's even concede that there's indeed a function. So what? That does not make them any less obvious as vestigial in regards to the original thing found in other species. The tail is here a fused set of bones. Yes, some muscles attach to it. It is still vestigial because it has the developmental pattern, the number of bones, etc, only the bones get fused and we get a tiny thing compared to the tail present in other organisms. The actual issue was not addressed, they used a distraction towards some function(s). But they don't deal with what the evidence actually is about.
If we pay no attention to whether they dealt with the actual issue we empower them. We make them think that they dealt with the issue all right, and then we have people desperately trying to prove that there's no function (a philosophical conundrum in itself), as if that was really important. Then we figure out a bit more, and there's a function for that particular pseudogene. It makes a regulatory RNA. Proteins bind to the region. Creationists come out of that thinking that they were right about all of it. Not a very productive day for science by either side. We ended up helping them out of not paying attention. We followed the red-herring.
Chris,
You are mistaken regarding HBBP conservation. The exon sequences are 1.45% divergent between human and chimp, while the intron sequences are 1.01% divergent. This suggests that any sequence conservation is in the introns, while the exons are evolving neutrally. (Those numbers are according to Moleirinho et al.) And this too contradicts the sequence conservation track on the UCSC genome browser, which shows the exons much more conserved than the introns. Odd.
And can I complain about Moleirinho et al. consistently saying "mutation rate" when they mean "fixation rate"?
Tomkins does not mention whether according to him HBBP1 makes any protein. It seems Tomkins just posits 'a function'. What function?
Tomkins argues from the scientific literature, based on statements as "some pseudogenes possess special functions, including regulating parental gene expression" ( Li, W., W. Yang, and X-J. Wang. 2013. Pseudogenes: Pseudo or real functional elements? Journal of Genetics and Genomics 40, no. 4: 171–177), and "cases of pseudogenes have been recently shown to act as endogenous competitive RNAs to their cognate genes" (Wen. Y. Z., L. L. Zheng, L. H. Qu, F. J. Ayala, and Z. R. Lun. 2012. Pseudogenes are not pseudo any more. RNA Biology 9, no. 1: 27–32.).
Some pseudogenes are transcribed, and their mRNA might compete with legitimate mRNA from the corresponding valid gene. It does not make sense to regard this as a case of gene regulation, and allot the pseudogene a regulatory function on that basis. It is noise in the system. Given the messy state of transcription, the valid gene has to get higher gene expression to drown the noise. Not only lack of function of the pseudogene, but disrupting remnant activity.
John, I posted your figure in an "UPDATE" above. Let me know if my explanation makes sense.
Larry, I don't think your explanation makes sense. Why are the pseudogene exons more conserved than the surrounding sequences, all of which diverged, presumably, at the same time? Conservation doesn't equate to sequence similarity but to greater similarity than expected from neutral evolution, and that's what the track would appear to show. Note also that the introns in all the globin genes appear to evolve significantly faster than the overall background rate (shown by the bars considerably below the line); I don't know what causes that either.
How did you get the track you posted?
So the whole β-globin locus is pretty much a mutational hot spot?
Whatever fits the paradigm...
It could very well be a mutational hot-spot for some or most of the non-random mutations via quantum coherence...
Larry,
After messing around with my track it seems to me that what taxa you include doesn't affect the graphic display; it shows conservation for 100 vertebrate species regardless. Looking at actual sequence, I find almost no difference among human, chimp, gorilla, and orangutan. Just as a quick visual impression, <1% divergence between any two of those species in exon sequences. I can't get the multiple alignment download to work, unfortunately; it seems stuck on a whole chromosome download rather than the limits I set.
Hello Larry
I finished my undergraduate studies in Biochemistry last year, and I recently waded into the turbulent rivers of the Evolution vs Creationism tussle. I accept the theory of evolution as a well-substantiated explanation of the diversity of life, but I need directions on how to deeply investigate the theory. I don't know whether you received any courses in evolution during your schooling, but I didn't so I would like to know how you began studying it, maybe it could guide me.
Finally, could you recommend good books on the fundamental and quite advanced concepts in evolution, I think it would help me know the facts, problems and proposed solutions to the problems facing evolutionary Theory.
Years ago I was looking at precisely this region, to make up a data example for use in a class. I made a tree (by parsimony, I think) and found that it looked fine, except Rhesus Macacque and human had switched positions. The apes were in their usual positions otherwise, except the sister group to chimps was Rhesus Macacque, and the human was the outgroup to all the apes. I was pretty sure that this was a data copying error at Genbank. Could this error still be there? I thought that it has since been corrected.
a mutational hot-spot for some or most of the non-random mutations via quantum coherence...
Holy moly. I hope that was intended as a joke.
Photosynthesis with its almost 100% efficient process of capturing light energy that hits the leaf was also considered a joke... until it was revealed that that plants use quantum coherence to maximize that efficiency...
Who says DNA molecules can't use the same process to maximize efficiency in DNA replication and minimize the spurious consequences of mutagenesis?
Who says DNA molecules can't use the same process to maximize efficiency in DNA replication and minimize the spurious consequences of mutagenesis?
Sure, just show me how DNA molecules achieve vibrational resonance with light energy. Of course that would *maximize* energy transfer and thus mutagenesis, not minimize it.
Do you want to stop now or embarrass yourself further?
Sure, just show me how DNA molecules achieve vibrational resonance with light energy. Of course that would *maximize* energy transfer and thus mutagenesis, not minimize it.
Lol.
You neither know what quantum coherence is nor how it works...
You don't have to continue to embarrass yourself...
This will do...lol
BTW: I was using photosynthesis process as an analogy. When I wrote the same process, I was referring to quantum coherence and not photosynthesis. Anybody who knew anything about quantum mechanics would have realized that... Unfortunately it is painfully obvious you don't know anything about it.
If I were you, I'd stop the embarrassment now...
@Joe
There’s clearly something wrong with the macaque sequence. You have to leave it out of any analysis or the results will be screwed up.
Offhand, I'd recommend Mark Kirkpatrick and Douglas Futuyma's Evolution, the third edition of the undergraduate evolutionary textbook originally written by Futuyma. Abotu $100 so not exactly cheap, but very respected and comprehensive.
You neither know what quantum coherence is nor how it works...
Actually, yep, I do. In photosynthesis in plants, the idea in these latest papers is, as I said, that it accounts for greater than classical efficiency of energy transfer.
Now if you want to speculate (since that’s what you are doing, speculating) there is something similar going on with DNA, first you need an energy source with frequencies that the electrons involved can vibrate harmonically with. This would be light in the photosynthesis example. Where’s your energy source with characteristic frequencies in the DNA example? Then even if you manage to find an energy source, the result of the coherence mechanism is greater energy. This doesn’t make DNA replication less error-prone. It either simply increases the rate of both normal replication and mutagenesis, or possibly, by imparting greater energy to the electrons involved, increases the probability of mutagenesis. Either way, more mutagenesis.
Sorry Jass, your speculation, even if true, would work exactly opposite of the way you want.
Some of you might be interested in this paper, published just two months ago in PNAS.
Duan, H.-G., Prokhorenko, V.I., Cogdell, R.J., Ashraf, K., Stevens, A.L., Thorwart, M., and Miller, R.D. (2017) Nature does not rely on long-lived electronic quantum coherence for photosynthetic energy transfer. Proceedings of the National Academy of Sciences 114:8493-8498. [doi: 10.1073/pnas.1702261114 ]
Abstract:During the first steps of photosynthesis, the energy of impinging solar photons is transformed into electronic excitation energy of the light-harvesting biomolecular complexes. The subsequent energy transfer to the reaction center is commonly rationalized in terms of excitons moving on a grid of biomolecular chromophores on typical timescales <100 fs. Today’s understanding of the energy transfer includes the fact that the excitons are delocalized over a few neighboring sites, but the role of quantum coherence is considered as irrelevant for the transfer dynamics because it typically decays within a few tens of femtoseconds. This orthodox picture of incoherent energy transfer between clusters of a few pigments sharing delocalized excitons has been challenged by ultrafast optical spectroscopy experiments with the Fenna–Matthews–Olson protein, in which interference oscillatory signals up to 1.5 ps were reported and interpreted as direct evidence of exceptionally long-lived electronic quantum coherence. Here, we show that the optical 2D photon echo spectra of this complex at ambient temperature in aqueous solution do not provide evidence of any long-lived electronic quantum coherence, but confirm the orthodox view of rapidly decaying electronic quantum coherence on a timescale of 60 fs. Our results can be considered as generic and give no hint that electronic quantum coherence plays any biofunctional role in real photoactive biomolecular complexes. Because in this structurally well-defined protein the distances between bacteriochlorophylls are comparable to those of other light-harvesting complexes, we anticipate that this finding is general and directly applies to even larger photoactive biomolecular complexes.
judmarc,
Actually, yep, I do. In photosynthesis in plants, the idea in these latest papers is, as I said, that it accounts for greater than classical efficiency of energy transfer.
You either don't know what you are talking about or you are contradicting yourself. Let's see which one it is. I suspect both:
The latest papers, that mentioned much greater efficiency of energy transfer in photosynthesis, almost without loss of energy to heat, much better than the classical energy transfer, what is that system of energy transfer called? lol
judmarc,
Where’s your energy source with characteristic frequencies in the DNA example? Then even if you manage to find an energy source, the result of the coherence mechanism is greater energy
It is the same source of energy that caused the spontaneous generation of life, and now looks like is behind the quantum coherent entanglement controlled mitosis, cell differentiation, non-random mutations and so on...
You should not be asking my this questions... You are the one who should know better what energy is behind OOL ;-)
Larry,
The study you refer to used only one model of experimentation available to see if Quantum Coherence is maintained long enough...
There have been many studies preformed before this and after that showed quantum coherence timescales well above 60 fs up 800 fs a and even more...
Bird navigation involving quantum coherence long-lived timescales to measure magnetic fields has been long disputed until recently...
Our sense of smell via quantum coherence as well as quantum consciousness via entanglement have been disputed for over 20 years until recent experiments have proven vibration in microtubules of neurons.
Photosynthesis is too efficient to use any conventional energy transfer system...just as judmarc admitted himself...Energy transfer via quantum coherence is way superior to the conventional one, just like quantum computers' efficiency is superior to the conventional ones...
No one can deny these facts... You either use it or lose it ;-)
Jass, I believe I see the problem (beyond your tendency to fall in love with science-y sounding words - this month, God apparently works mysteriously via quantum coherence, his wonders to perform): A complete lack of reading comprehension. In your October 23rd comment, you first say I don’t know what I’m talking about, then you entirely agree with me (about the way the recent papers say quantum coherence works in photosynthesis).
It is the same source of energy that caused the spontaneous generation of life
This was Jass’s response when asked about the source of energy at particular frequencies (i.e., radiant energy) that would operate for DNA replication as light does in photosynthesis.
So tell us, Jass, what is God’s frequency?
(You also never gave an answer to my objection that higher energy transfer in DNA synthesis would lead to greater, not lesser, mutagenesis. Why are people advised to wear sunscreen, Jass? Think about it.)
Thanks Larry. Not unusual that there would be a potentially exciting finding that is later determined not to be the case. Jass will have to find some other machina for his Deus.
just like quantum computers' efficiency is superior to the conventional ones.
Specifically for what types of problems? I.e., for what classes of mathematical problems do there exist mathematical proofs of the superiority of quantum computing? For what classes are there mathematical proofs that there is no "quantum speedup"? And what are some of the classes for which there are as yet no proofs either way? Since you used this as an example, Jass, I'm sure you know about it.
@judmarcWednesday,
Thanks Larry. Not unusual that there would be a potentially exciting finding that is later determined not to be the case. Jass will have to find some other machina for his Deus.
That's why so many experiments need to be repeated especially when study shows conflicting results with so many other experiments...
Coherence in Photosynthesis
"It is unclear how energy absorbed by pigments in antenna proteins is transferred to the central site of chemical catalysis during photosynthesis. Hildner et al. (p. 1448) observed coherence—prolonged persistence of a quantum mechanical phase relationship—at the single-molecule level in light-harvesting complexes from purple bacteria. The results bolster conclusions from past ensemble measurements that coherence plays a pivotal role in photosynthetic energy transfer. Hayes et al. (p. 1431, published online 18 April) examined a series of small molecules comprised of bridged chromophores that also manifest prolonged coherence.
Abstract
The initial steps of photosynthesis comprise the absorption of sunlight by pigment-protein antenna complexes followed by rapid and highly efficient funneling of excitation energy to a reaction center. In these transport processes, signatures of unexpectedly long-lived coherences have emerged in two-dimensional ensemble spectra of various light-harvesting complexes. Here, we demonstrate ultrafast quantum coherent energy transfer within individual antenna complexes of a purple bacterium under physiological conditions. We find that quantum coherences between electronically coupled energy eigenstates persist at least 400 femtoseconds and that distinct energy-transfer pathways that change with time can be identified in each complex. Our data suggest that long-lived quantum coherence renders energy transfer in photosynthetic systems robust in the presence of disorder, which is a prerequisite for efficient light harvesting.
There are few more...
judmarc,
Specifically for what types of problems? I.e., for what classes of mathematical problems do there exist mathematical proofs of the superiority of quantum computing? For what classes are there mathematical proofs that there is no "quantum speedup"? And what are some of the classes for which there are as yet no proofs either way? Since you used this as an example, Jass, I'm sure you know about it.
Quantum vs Classical Computation
"Quantum Computing is the art of using all the possibilities that the laws of quantum mechanics give us to solve computational problems. Conventional, or "Classical" computers (like the one used to build this page) only use a small subset of these possibilities. In essence, they compute in the same way that people compute by hand. There are many results about the wonderful things we would be able to do if only we had a large enough quantum computer. The most important of these is probably that we would be able to perform simulations of quantum mechanical processes in physics, chemistry and biology, which will never come within the range of classical computers. Let's compare some aspects of classical and quantum computers:
http://www.thphys.nuim.ie/staff/joost/TQM/QvC.html
judmarc,
So tell us, Jass, what is God’s frequency?
So tell us, judmarc, what is your dumb luck's source of creative energy that lead from lifeless matter becoming alive and how intelligent people like yourself replicate what dumb luck accomplished? It should be a piece of cake for clever guys like you lol
Please feel free to include the frequencies your god dumb luck operated on when it converted lifeless matter into life... Don't overwhelm us with too much evidence!
Few experimental evidence will do... ;-)
I keep my fingers crossed for one piece of evidence...lol
Birds measure magnetic fields using long-lived quantum coherence
http://physicsworld.com/cws/article/news/2016/apr/07/birds-measure-magnetic-fields-using-long-lived-quantum-coherence
A system like that must have evolved! By what mechanism? It remains to be seen in the quantum realm of subatomic particles... ;-)
One more on quantum coherence in photosynthesis from Larry's neck of the woods U of T...
Coherently wired light-harvesting in photosynthetic marine algae at ambient temperature
Elisabetta Collini1,3,4, Cathy Y. Wong1,3, Krystyna E. Wilk2, Paul M. G. Curmi2, Paul Brumer1 & Gregory D. Scholes1
Department of Chemistry, Institute for Optical Sciences and Centre for Quantum Information and Quantum Control, University of Toronto, 80 St George Street, Toronto, Ontario, M5S 3H6 Canada
School of Physics and Centre for Applied Medical Research, St Vincent’s Hospital, The University of New South Wales, Sydney, New South Wales 2052, Australia
These authors contributed equally to this work.
Present address: Dipartimento di Scienze Chimiche, Università di Padova, via Marzolo 1, 35100, Padova, Italy.
Correspondence to: Gregory D. Scholes1 Correspondence and requests for materials should be addressed to G.D.S. (Email: gscholes@chem.utoronto.ca).
Photosynthesis makes use of sunlight to convert carbon dioxide into useful biomass and is vital for life on Earth. Crucial components for the photosynthetic process are antenna proteins, which absorb light and transmit the resultant excitation energy between molecules to a reaction centre. The efficiency of these electronic energy transfers has inspired much work on antenna proteins isolated from photosynthetic organisms to uncover the basic mechanisms at play1, 2, 3, 4, 5. Intriguingly, recent work has documented6, 7, 8 that light-absorbing molecules in some photosynthetic proteins capture and transfer energy according to quantum-mechanical probability laws instead of classical laws9 at temperatures up to 180 K. This contrasts with the long-held view that long-range quantum coherence between molecules cannot be sustained in complex biological systems, even at low temperatures. Here we present two-dimensional photon echo spectroscopy10, 11, 12, 13 measurements on two evolutionarily related light-harvesting proteins isolated from marine cryptophyte algae, which reveal exceptionally long-lasting excitation oscillations with distinct correlations and anti-correlations even at ambient temperature. These observations provide compelling evidence for quantum-coherent sharing of electronic excitation across the 5-nm-wide proteins under biologically relevant conditions, suggesting that distant molecules within the photosynthetic proteins are ‘wired’ together by quantum coherence for more efficient light-harvesting in cryptophyte marine algae.
So neither any of my questions nor my objection have been answered. Not that anything else was expected.
You know, when someone asks you a question like what classes of mathematical problems have mathematical proofs showing them to be capable of quantum speedup, rather than quoting generic pablum it's OK to say you don't know the specifics.
So I looked at the bird article. The scientists there feel they've built a more realistic model. They are quite a long way from proving anything, but perhaps they'll be able to accumulate actual evidence in support of their model in the coming years. It will be interesting to see if they can.
But why it should be any surprise to you that all of chemistry, including biochemistry, works in accordance with the laws of quantum physics, I really don't know. Since the universe works in accordance with these laws, what else would you expect?
Quantum Biometrics Exploits the Human Eye’s Ability to Detect Single Photons
Identifying individuals by the way their eyes detect photons could be a hugely accurate form of biometrics, guaranteed by the laws of quantum mechanics.
https://www.technologyreview.com/s/604266/quantum-biometrics-exploits-the-human-eyes-ability-to-detect-single-photons/
This quantum feature of human eye must have evolved mainly by non-random mutations and random natural selection... which some of which are also directed by quantum mechanics... That's something evolution would predict...I think...
judmarc,
They are building quantum solar panels imitating the quantum coherence process of photosynthesis because it is 100% efficient. You are stuck in 17 century Newtonian physics... It's better for me because I'm thinking about investing... ;-) You do your own thing!
"Unlocking nature's quantum engineering for efficient solar energy
Unlocking nature’s quantum engineering for efficient solar energy
(Phys.org)—Quantum scale photosynthesis in biological systems which inhabit extreme environments could hold key to new designs for solar energy and nanoscale devices. Certain biological systems living in low light environments have unique protein structures for photosynthesis that use quantum dynamics to convert 100% of absorbed light into electrical charge, displaying astonishing efficiency that could lead to new understanding of renewable solar energy, suggests research published today in the journal Nature Physics.
Read more at: https://phys.org/news/2013-01-nature-quantum-efficient-solar-energy.html#jCp
My goodness can't you just shut up for a moment with all your quantum-woo bullshit?
Apologies for my role in pointing out the idiocy and thus fostering more of it, though thanks to Larry for the reference to interesting scientific work on the topic.
off topic, but oh wow
https://www.nature.com/articles/ncomms13924.pdf
any chance a thread can be made of this?
@Mikkel Rumraket Rasmussen,
My goodness can't you just shut up for a moment with all your quantum-woo bullshit?
You're angry? I'd thought you would be happy that quantum mechanics is leading to another layer of sophistication in lifesystems clearly pointing toward God...
Wait until you see proof what QM does in DNA, non-random mutations, "pseudogenes", overlapping sequences for protein coding, transcription factor binding, dual coding genes, and much, much more...
Isn't this exciting?
Jass,
I should not be feeding your trolling, but seems like you're a bit confused.
"You're angry? I'd thought you would be happy that quantum mechanics is leading to another layer of sophistication in lifesystems clearly pointing toward God..."
Quantum mechanics is a model for physical phenomena as observed at very small levels. It applies to all physical phenomena, only, being about the extremely small, putting those tiny details together to try and explain things at a higher level requires lots of computation. If you want to call that "sophisticated," that's all right. The point is: it's a model for all physics. Physics, I hope you know, is natural.
So, I really don't understand how would anybody jump from: scientists are trying to explain some biological phenomena using quantum mechanics, therefore gods.
It's like being surprised that, after suggestion of the composition of matter out of atoms, you were surprised that life is also composed of atoms. Therefore gods.
The difficulty in the computation is a human problem, by the way. Nature keeps going regardless of our mathematical models being "sophisticated" or simple. It's still nature.
Hopefully now you can stop the useless and nonsensical bullshitting. Given your many posts above, I doubt that you're able to grasp the message, but there you go. This is it from me. I'd rather not continue. After all, this is off-topic.
@Gabriel Moreno-Hagelsieb,
I should not be feeding your trolling...
Now you know how I feel...
I tumbled onto this thread as a consequence of Christine Janis calling my attention to Jonathan's mentioning my "Evolution Slam Dunk" in his Panda's Piece. Since Tomkins is at top of YEC fact claimant food chain on genetic matters, this posting or Larry's will be added to my #TIP dataset www.tortucan.wordpress.com along with the various back and forths.
My general #TIP point is that it's always good to engage antievolutionists at the source documentation level, first because it allows the best of targeted criticism to be made, and second because in the process you end up clarifying and strengthening one's own understanding of the issues. Win/win.
Jass said:
"just like quantum computers' efficiency is superior to the conventional ones."
I work in Quantum computers and this isn't true either.
Post a Comment