This jumble of different kinds of genes makes it difficult to estimate the total number of genes in the human genome. The current estimates are about 20,000 protein-coding genes and about 5,000 genes for functional RNAs.
Aside from the obvious highly conserved genes for ubiquitous RNAs (rRNA, tRNAs etc.), protein-coding genes are the easiest to recognize from looking at a genome sequence. If the protein is expressed in many different species then the exon sequences will be conserved and it's easy for a computer program to identify the gene. The tough part comes when the algorithm predicts a new protein-coding gene based on an open reading frame spanning several presumed exons. Is it a real gene?
SUMMARY
There are about 20,000 potential protein-coding genes in the human genome. Proteins for 85% of these genes have been detected by various assays.Over the years, the number of protein-coding genes in the human genome has dropped from the initial reports of about 30,000 to about 20,000. All these estimates were in line with the predictions of knowledgeable exerts based on data going back to the 1960s.1
This drop in the number of predicted genes is fairly typical of genome studies. The gene-finding algorithms tend to over-predict the number of protein-coding genes and subsequent annotation leads to a "finished" genome sequence that eliminates all the false positives. This is one reason why you shouldn't trust the initial estimates of gene number in newly published genomes. Most of them will never be "finished" so the number of genes will always be inflated by a large number of false positives. Many of these unconfirmed genes will be ORFans—possible genes that are unique to that species. Don't get excited about ORFans unless you are dealing with a finished, well-annotated, genome.
The most obvious way of confirming a potential protein-coding gene is to find and confirm synthesis of a functional protein. Hundreds of human genes have been intensely studied over the years and in those cases there's no doubt at all about their existence. Several thousand more genes have matched to proteins in various ways.
That still left thousand of putative genes with no evidence that they actually make a protein.
Advancing technology in the past 15 years has made it possible to identify individual proteins even though they may be present in just a few copies per cell. The technique relies on mass-spectrometry and the existence of a well-annotated genome sequence [Genomics, Proteomics and Mass Spectrometry] [Biochemistry and Mass Spectrometry].
Here's a brief description of how it works. Take any tissue or groups of cells and isolate all the proteins. Digest them with a protease—enzymes that chop the protein into small pieces by cutting at a specific site. Trypsin is commonly used in these studies; it catalyzes cleavage of the peptide bond on the carbonyl side of lysine (K) and arginine (R) residues producing peptide fragments that all end with lysine and arginine.
The next step involves "weighing" all of the thousands of peptides in a mass spectrometer. You can visualize the process by imagining that the mixture of peptides is sprayed into one end of a long tube surrounded by an electromagnetic field that makes the charged peptides "fly" to the far end of the tube.2 Small peptides fly faster than larger ones so if you measure the time of flight (TOF) you can calculate the mass of the peptides. That's why it's called "mass spectrometry."
At the end of the experiment you have a huge list of the molecular weights of thousands and thousands of individual peptides. Here comes the fun part.
If you look at the coding sequence of a gene you can predict all of the peptides that will be produced by digesting with various proteases. For example, one of the predicted fragments of the human serum albumin protein is FKDLGEENFK (ends in lysine=K). This has an exact molecular mass of 1226.59 so if you find a peptide of that size in your mixture then it probably means that human serum albumin was present. In practice you want to identify several unique peptide masses for each protein just to be sure.
This technique only works if you have a reliable gene sequence and it only works on the genome level if you have a "finished" genome sequence, properly annotated so you can identify all potential reading frames. It only works if you have good computer programs and lots of memory and storage. It only works is you have very good mass spectrometers and experts who know how to use them. We do a simple experiment like this in our undergraduate biochemistry lab (3rd year) using a purified protein from a bacterial species with a small genome. It doesn't always work. That's several orders of magnitude easier than identifying all the proteins in a species with a large complex genome.
All those things can be found at several top-notch locations around the world. Recently two groups have published the results of massive mass-spec analyses of human proteins from many different tissues in order to determine the full extent of the human proteome.
Kim, M.-S., Pinto, S.M., Getnet, D., Nirujogi, R.S., Manda, S.S., Chaerkady, R., Madugundu, A.K., Kelkar, D.S., Isserlin, R., Jain, S., Thomas, J.K., Muthusamy, B., Leal-Rojas, P., Kumar, P., Sahasrabuddhe, N.A., Balakrishnan, L., Advani, J., George, B., Renuse, S., Selvan, L.D.N., Patil, A.H., Nanjappa, V., Radhakrishnan, A., Prasad, S., Subbannayya, T., Raju, R., Kumar, M., Sreenivasamurthy, S.K., Marimuthu, A., Sathe, G.J., Chavan, S., Datta, K.K., Subbannayya, Y., Sahu, A., Yelamanchi, S.D., Jayaram, S., Rajagopalan, P., Sharma, J., Murthy, K.R., Syed, N., Goel, R., Khan, A.A., Ahmad, S., Dey, G., Mudgal, K., Chatterjee, A., Huang, T.-C., Zhong, J., Wu, X., Shaw, P.G., Freed, D., Zahari, M.S., Mukherjee, K.K., Shankar, S., Mahadevan, A., Lam, H., Mitchell, C.J., Shankar, S.K., Satishchandra, P., Schroeder, J.T., Sirdeshmukh, R., Maitra, A., Leach, S.D., Drake, C.G., Halushka, M.K., Prasad, T.S.K., Hruban, R.H., Kerr, C.L., Bader, G.D., Iacobuzio-Donahue, C.A., Gowda, H., and Pandey, A. (2014) A draft map of the human proteome. Nature, 509:575-581. [doi: 10.1038/nature13302]Kim et al. (2015) looked at 30 different types of cells and tissues and identified proteins encoded by a total of 17,294 protein-coding genes. Of these, 2,535 represented genes for which no protein have previously been identified.
Wilhelm, M., Schlegl, J., Hahne, H., Gholami, A.M., Lieberenz, M., Savitski, M.M., Ziegler, E., Butzmann, L., Gessulat, S., Marx, H., Mathieson, T., Lemeer, S., Schnatbaum, K., Reimer, U., Wenschuh, H., Mollenhauer, M., Slotta-Huspenina, J., Boese, J.-H., Bantscheff, M., Gerstmair, A., Faerber, F., and Kuster, B. (2014) Mass-spectrometry-based draft of the human proteome. Nature, 509:582-587. [doi: 10.1038/nature13319]
About 2,400 genes are "housekeeping" genes whose protein products are found in all cells. About 1,500 genes were only expressed in one of the 30 tissues and cells types analyzed.
According to Kim et al. there are still several thousand potential protein-coding genes that have not been confirmed by detecting a protein product. In addition, they found 44 new ORFs that have not been annotated in the latest release of the human genome. These are potential new genes but the authors caution that the proteins may not have a function. Only 144 pseudogenes produced a polypeptide out of about 15,000 in the human genome. This is not unexpected since recently inactivated genes might still produce nonfunctional protein or protein fragments. It reminds us that cells can produce junk proteins as well as junk DNA.
The other group looked at mass-spec data that had been published in the past ten years. A total of from 27 different tissues were examined(Wilhelm et al, 2015). They constructed a database (ProteomicsDB) that accounted for 18,097 protein-coding genes out of the total of 19,629 that were annotated in the Swiss-Prot database. This accounts for almost all of the potential protein-coding genes. (Some genes might be expressed in very restricted cells at very limited times during development. Other genes produce proteins that can't be detected by the techniques used in most studies. Other "genes" might actually be pseudogenes.)
Wilhelm et al. estimate that the human genome contains 10,000-12,000 core genes that are ubiquitously expressed (= "housekeeping" genes).
These authors also looked at potential coding regions in large non-coding RNAs (lncRNAs). There are more than 21,000 potentially functional lncRNAs in the human genome although almost nobody believes that they are all functional (but see 3,000 new genes discovered in the human genome - dark matter revealed ]. Wilhelm et al. found that 404 of these potential lncRNA genes encode detectable peptides. Here's what they say about them ...
"The biological significance of translated lincRNAs and [other RNAs] is not clear at present. These may constitute proteins 'in evolution' representing hitherto undiscovered biology or arise by stochastic chance marking such proetins as 'biological noise.'"The results from these two groups indicate that we know almost all the protein-coding genes in the human genome (~20,000) and there are likely to be very few undiscovered protein-coding genes.
Some scientists are skeptical. Ezkurdia et al. (2014) think that both studies over-estimate the number of genes that are expressed. Their arguments are convincing but I don't think it makes a huge difference.
The skeptical scientists are worried about the conflict between the results in the two Nature papers and the results from a huge international effort called the Chromosome-Centric Human Proteome Project (C-HPP). The C-HPP Consortium has just published a bunch of papers in the September (2015) issue of the Journal of Proteome Research.
Their new database incorporates the data from the two Nature papers (Kim et al., 2014; Wilhelm et al., 2015) and comes up with different numbers of confirmed proteins. The Human Proteome Project (HPP) is an extension of C-HPP. Their most recent database has 16,491 "confident" proteins based on mass-spec and other types of experiments (Omenn et al., 2015). The pie-chart3 below summarizes the data.
You can see that almost all of the "confirmed" proteins have been detected by mass-spec. There are still about 3,000 potential protein-coding genes that have not been confirmed.
The lead paper in the journal summarizes all of the work and highlights two papers that discuss ways of identifying new genes (Paik et al., 2015). They issue this caution that's undoubtedly directed at workers outside of the C-HPP Consortium.
These two papers provide a thorough examination of the challenges of claiming, confirming, and validating peptide findings and protein matches. We recommend that all investigators scrutinize the discussion sections of these paper and apply the guidelines to their own data sets and other publicly available data sets. Such quality assurance will be subjected to open discussion at the HUPO 2015 Congress in Vancouver. Claims of novel translated products from pseudogenes or long noncoding RNAs require at least as great scrutiny as missing proteins from genes with transcripts or homologies, including use of class-specific FDRs.
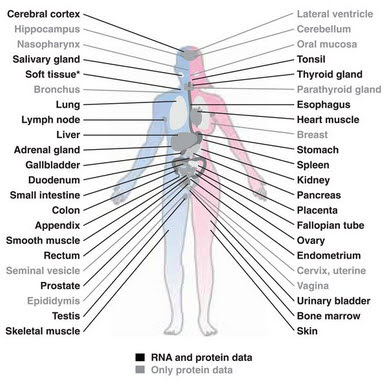
Then there's the Human Protein Atlas program. The results are summarized in a Science paper from last January (Uhlén et al., 2015). This project examined 44 different tissues and compiled data on RNA expression, proteins expression, and tissue localization using more than 20,000 antibodies.
They identified proteins from 17,132 genes out of 20,344 (84%) in at least one study but only 13,841 that were confirmed in at least two studies. Of these, 8,874 (44%) were detected in all tissues. These housekeeping genes encode mitochondrial proteins, basic metabolism proteins, structural proteins including many membrane proteins, and proteins required for transcription, translation, DNA replication, and processing/modification.
Once again, the bottom line is that most of the potential protein-coding genes have been confirmed. There's still some doubt about some of the putative protein-coding genes that have no supporting evidence. The human genome contains at least 17,000-18,000 protein-coding genes but probably no more than 21,000.
1. There's a widespread myth going around that the experts were "shocked" to discover only 30,000 genes when the human genome sequence was completed. See False History and the Number of Genes, and Facts and Myths Concerning the Historical Estimates of the Number of Genes in the Human Genome.
2. This "electrospray mass spectrometry" has now been mostly replaced by "matrix-assisted laser desorption ionization" or MALDI. When coupled to time of flight measurements the technique is known as MADLI-TOF. The details of the technique aren't important.
3. Don't forget that my wife's direct ancestor, William Playfair, invented pie charts! [Bar Graphs, Pie Charts, and Darwin]
Ezkurdia, I., Vázquez, J., Valencia, A., and Tress, M. (2014) Analyzing the First Drafts of the Human Proteome. Journal of Proteome Research, 13(8), 3854-3855. [doi: 10.1021/pr500572z]
Omenn, G. S., Lane, L., Lundberg, E. K., Beavis, R. C., Nesvizhskii, A. I., and Deutsch, E. W. (2015) Metrics for the Human Proteome Project 2015: Progress on the Human Proteome and Guidelines for High-Confidence Protein Identification. Journal of Proteome Research, 14(9), 3452-3460. [doi: 10.1021/acs.jproteome.5b00499]
Uhlén, M., Fagerberg, L., Hallström, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., Sivertsson, Å., Kampf, C., Sjöstedt, E., and Asplund, A. (2015) Tissue-based map of the human proteome. Science, 347(6220), 1260419. [doi: 10.1126/science.1260419 ]





22 comments :
You forgot to tell us how this confirms Intelligent Design? :-)
This type of good factual summary of recent papers of interest is very much appreciated, though due to lack of pointless name calling and specious argument it is very unlikely to attract as many comments as some of your other posts.
How 'bout I add some pointless name calling and specious arguments to the comments? Would that work?
In all seriousness why is the ID conversation so much more attractive to your readers? In October over %75 of the comments on your posts were made on ID related articles. Why aren't the scientists that read your blog actually interested in the actual science? To be fair you do have a few posts that aren't science or ID that are treated the same. Why does ID drive such interest?
Beau,
When Larry switches back to science, Barry think he's won:
After two failed posts, Larry has put up a post on a completely unrelated topic, apparently giving up on even a pretense of backing up his claim. I expect to see him post an apology for his smear against me that, when challenged, he was unable to support (as soon as pigs fly).
Personally, I much prefer science to ID. ID is OK for occasional entertainment, but too much concentrated idiocy in a short span of time has a nauseating effect on me.
Beau Stoddard asks:
In all seriousness why is the ID conversation so much more attractive to your readers? In October over %75 of the comments on your posts were made on ID related articles. Why aren't the scientists that read your blog actually interested in the actual science? To be fair you do have a few posts that aren't science or ID that are treated the same. Why does ID drive such interest?
I think the interest is sociological.
At least two reasons, I think.
1) Comments are often in response to cdesign proponentsists' comments, and IDiots are less likely to enter into pure science posts.
2) Commenters often feel they have less to add to the science posts, and commenters to these are usually those with special expertise in just that field; thus the commenter pool is reduced.
Beau,
ID is an important topic. The majority of people in this world are predisposed toward “wanting to believe” quite unsupported things. But it is becoming increasingly difficult to maintain belief in typical religious doctrines and dogma in the face of increasing scientific understanding of the nature of things. So, along comes ID which claims a scientific basis for god.
Note that ID, even if true, would have no bearing on matters such as the existence of heaven and hell, or a lord that loves us and is in control, or the wishful thinking that death is not the end, or the efficacy of prayer, etc. But that doesn’t matter to someone who is predisposed to wanting to believe these things.
ID arose for political reasons, but for the average person it merely has the potential to justify believing in things that are hoped for, but for which there is no evidence. Many of us on this site are not so much opposed to religious practise in its most benign forms, but rather the irrationality that attends religious belief of all sorts. Sam Harris once wrote (paraphrasing here): if you examine the origins of every human atrocity you will find each and every time that the promoters and perpetrators of that atrocity were motivated by the belief in absurd, untrue things. Irrationality is the problem and falsely claiming there is a scientific justification for any irrational belief system only helps to maintain a human population that is dangerously credulous.
I'm trying to make sense out of this amazing post with my ultra limited knowledge, but there's a couple of things I'm not sure about
About 2,400 hundred genes are "housekeeping" genes
Is that 2400 or 240000 please? If there are 20000 genes I guess it's the former right?
It reminds us that cells can produce junk proteins as well as junk DNA
Would that be junk RNA?
Oh, 20000 is the number of protein coding genes, if the genome is 3000M bases long and about a 10% are genes, that's 300M genes, so 240K would be entirely plausible for housekeeping genes I guess?
Larry or others: regarding the finding that 144 out of 15,000 pseudogenes produce products, it is a small percentage (0.01%). The number of products that will be functional (for the reason you gave in post) will be low. Are their any reliable estimates/speculations on the fraction that could be functional in some way?
No, your math and figuring is all wrong. First, if 10% of the 3000 million nucleotide genome is devoted to genes, that is 300 mllion nucleotides - but each nucleotide is not a gene. Second, if the total number of genes is 20,000-25,000 or so, then the fraction that would be "housekeeping genes" would be a much smaller number.. quite possibly in the order of 2,000 to 3,000 or so.
Thanks SRM, I certainly need to learn the basics. A quick google search showed that genes are (or can be?) thousands of bp's long.
So I take it "2,400 hundred genes are "housekeeping" genes" should be corrected to "2,400 genes are "housekeeping" genes"?
Oh, I didn't notice the 2,400 hundred error in the post. Yes, I presume the number should be simply 2,400. However, as the post relates some investigators claim that a larger fraction of the 20,000 or so protein coding genes could be considered "housekeeping" (i.e. essential for the basic functions of the cell) but all of this is based on expression patterns rather than detailed knowledge of function, I think.
Yes, I believe the average bacterial gene is about 1000 bp long. The average eukaryotic gene (in terms of amino acid coding regions, i.e, exons) is only slightly larger but with non-coding introns included the gene may be many 10s of thousands of bp long.
Commenters often feel they have less to add to the science posts, and commenters to these are usually those with special expertise in just that field; thus the commenter pool is reduced.
Yes. I read the science-oriented posts just as much as the ones about superstitious beliefs like religion and creationism. But on the former I just shut up an listen to the experts. There's not much for me to add.
On another board I frequent, I've noticed that many of the most protracted discussions tend to involve one or two recalcitrant denialists and the attempts of others to educate them. I recall one thread that went on for about 130 pages over 8 months in which a Muslim who, based on his reading of the Quran, was convinced that the earth did not revolve or rotate resisted all efforts to explain why this was false. They dynamic is of informed people trying their best to cure someone of his ignorance, when that person is highly invested in remaining ignorant.
Larry, this is what I and others need to know more about but it's stuck behind a paywall and the article sure does not explain much:
Complex grammar of the genomic language
http://www.kurzweilai.net/forums/topic/complex-grammar-of-the-genomic-language
Two or them have PubMed Central versions available for free;
Proteomics. Tissue-based map of the human proteome
http://www.ncbi.nlm.nih.gov/pubmed/25613900
Analyzing the First Drafts of the Human Proteome
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4334283/
Beau:
Why aren't the scientists that read your blog actually interested in the actual science? To be fair you do have a few posts that aren't science or ID that are treated the same. Why does ID drive such interest?
Just as any fool can be an ID proponent, any fool can point out their errors. Peer-reviewed science is naturally less likely to attract comment, even from those in the field.
Beau - Google "bikeshedding."
Thanks for the links Jim. It's good for me to stay current in what is available for data.
What I could not find though is the spatial and behavioral information needed to begin to model human cell nuclei. Behavioral information includes TF to TF interactions, as in a thread I linked to. Spatial information would show the 3D arrangement of normal uncoiled chromosomes, the "territories".
Please let me know of any information you know of that gets into the functional details needed to model at least a portion of a nucleus. With the way information is now scattered around what is needed might already exist at a place that does not get as much news coverage.
Some other interesting news:
TSRI and St. Jude Scientists Help Launch Human Dark Proteome Initiative
https://darkproteome.wordpress.com/about/the-dark-proteome-animated/
http://www.scripps.edu/newsandviews/i_20151116/dark_proteome.html
I am not sure yet what methods are they gonna use to estimate which proteins are disordered.
I'm not sure this post addresses the original question, which was, "How many proteins do humans make?" There are 17-18K protein coding genes. If one assumes one gene, one protein, then I guess it implies there are 17-18K proteins. But given alternative splicing, post-translational modifications, etc., there can be many proteins associated with a given protein-coding gene. In fact, some genes seem to encode 1000s of different proteins. So if there are 17-18K protein coding genes, humans make many more than this. This so far doesn't directly answer the question either but does set a lower limit. So the question stands: How many proteins do humans make? Would be curious to hear your thoughts.
Post a Comment