This isn't good enough for many human chauvinists. They are still looking for something special that sets human apart from all other animals. I listed seven possibilities in my post on the deflated ego problem:
You can see how many of these positions are related to ongoing controversies in molecular biology, especially the debate over junk DNA. Keep in mind that what's behind that debate (junk DNA) is not only differing views about the strength and importance of natural selection but also a sense on behalf of some scientists that the "specialness" of humans requires a special explanation.1. Alternative Splicing: We may not have many more genes than a fruit fly but our genes can be rearranged in many different ways and this accounts for why we are much more complex. We have only 25,000 genes but through the magic of alternative splicing we can make 100,000 different proteins. That makes us almost ten times more complex than a fruit fly. (Assuming they don't do alternative splicing.)2. Small RNAs: Scientists have miscalculated the number of genes by focusing only on protein encoding genes. Our genome actually contains tens of thousands of genes for small regulatory RNAs. These small RNA molecules combine in very complex ways to control the expression of the more traditional genes. This extra layer of complexity, not found in simple organisms, is what explains the Deflated Ego Problem.3. Pseudogenes: The human genome contains thousands of apparently inactive genes called pseudogenes. Many of these genes are not extinct genes, as is commonly believed. Instead, they are genes-in-waiting. The complexity of humans is explained by invoking ways of tapping into this reserve to create new genes very quickly.4. Transposons: The human genome is full of transposons but most scientists ignore them and don't count them in the number of genes. However, transposons are constantly jumping around in the genome and when they land next to a gene they can change it or cause it to be expressed differently. This vast pool of transposons makes our genome much more complicated than that of the simple species. This genome complexity is what's responsible for making humans more complex.5. Regulatory Sequences: The human genome is huge compared to those of the simple species. All this extra DNA is due to increases in the number of regulatory sequences that control gene expression. We don't have many more protein-encoding regions but we have a much more complex system of regulating the expression of proteins. Thus, the fact that we are more complex than a fruit fly is not due to more genes but to more complex systems of regulation.6. The Unspecified Anti-Junk Argument: We don't know exactly how to explain the Deflated Ego Problem but it must have something to do with so-called "junk" DNA. There's more and more evidence that junk DNA has a function. It's almost certain that there's something hidden in the extra-genic DNA that will explain our complexity. We'll find it eventually.7. Post-translational Modification: Proteins can be extensively modified in various ways after they are synthesized. The modifications, such as phosphorylation, glycosylation, editing, etc., give rise to variants with different functions. In this way, the 25,000 primary protein products can actually be modified to make a set of enzymes with several hundred thousand different functions. That explains why we are so much more complicated than worms even though we have similar numbers of genes.
The latest contribution is in a recent issue of Nature that contains a series of "Insight" reviews on "Transcription and Epigenetics." The review I want to discuss is by de Laat and Duboule (2013). The abstract outlines a new hypothesis concerning the evolution of high-order chromatin structure.
How a complex animal can arise from a fertilized egg is one of the oldest and most fascinating questions of biology, the answer to which is encoded in the genome. Body shape and organ development, and their integration into a functional organism all depend on the precise expression of genes in space and time. The orchestration of transcription relies mostly on surrounding control sequences such as enhancers, millions of which form complex regulatory landscapes in the non-coding genome. Recent research shows that high-order chromosome structures make an important contribution to enhancer functionality by triggering their physical interactions with target genes.The opening paragraph (below) makes it clear that they are discussing the deflated ego problem.
Access to animal genome sequences has revealed that the level of complexity of an organism does not relate to its number of genes. Mammals are more complex in morphology and behaviour than roundworms, but their genomes both contain around 20,000 genes. Various parameters can contribute to increased complexity, such as the extent of protein modifications or the diversity of splicing patterns. Pleiotropy is another possible contributor, whereby genes acquire multiple functional tasks at different times and places either during development or in adult life. In this case, gene regulation, rather than function, had to evolve to associate regulatory alternatives to particular genes. Although gene transcription is initiated at promoters, which recruit the basal transcription machinery, these sequences have little impact on transcription control during development and hence this latter task mostly relies on enhancers.The authors claim that there are millions of enhancers in the human genome. If we take "millions" to mean just two million then there are, on average, one hundred enhancers per gene. This means that expression of each gene in our genome is regulated, on average, by the binding of 100 transcription factors to 100 transcription factor binding sites (= enhancers). Thus, a lot of our genome (40%) is devoted to regulation.
Enhancers are sequence modules that contain binding motifs for transcription factors. They are preferentially located in the non-coding part of the genome, at various distances from their target genes. In mammals, more than 95% of the genome is non-coding and large gene deserts can sometimes span several megabases. The recent development of high-throughput methods has made it possible to systematically search for enhancers; millions of such regulatory modules have been predicted, with 40% of our genome now estimated to carry some regulatory potential.So far, there's nothing that makes humans, or vertebrates, special. After all, fruit flies and roundworms may also have 100 enhancers per gene. Here's where it gets interesting because the authors are proposing that vertebrates (mammals?) have evolved something special.
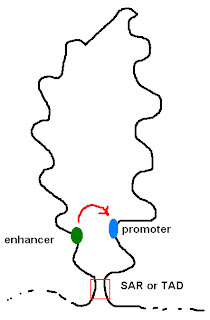
Evolution of mammalian enhancer landscapesThe authors illustrate their claim with the figure shown below. I've modified it to focus on the main point; namely, that mammals have evolved the special ability to control gene expression using many transcription factors that can function at great distances from the promoter.
Vertebrate genomes are unique in that they contain large gene deserts with enhancers acting over distances in the megabase range (see ref. 9 for a review). Invertebrate species studied so far tend to have more local regulatory controls, which can often be recapitulated by short transgenes, such has been shown for the roundworm Caenorhabditis elegans. Admittedly, in Drosophila, gene regulation during development is complex, with multiple enhancers acting on individual genes and some loci controlled by series of intricate enhancers. However, these enhancer–promoter interactions generally occur over distances shorter than 50 kb.
The diagram is a bit deceptive because it's not to scale. A typical mammalian gene has about 2000bp (2kb) of coding region (~exons1). It includes about 20kb of intron sequences.2 The authors are suggesting that expression of these genes is regulated by 100 enhancers that can act on the promoter at distances of more than 1000kb (1Mb). Bound transcription factors can contact the transcription initiation complex (i.e. RNA polymerase holoenzyme) at the promoter by forming a large loop of DNA. According to the review, the average lop size in mammals is 120kb or six times the size of a typical gene. The biggest loop discovered so far is 1,300kb.
It's true that genomes (prokaryotic and eukaryotic) are organized into large loops formed by proteins binding at Scaffold Attachment Regions (SARs) or Topological Associating Domains (TADs). It's true that a transcription factor could bind at an enhancer near the base of the loop and a promoter could be found at the base of the other side of the loop. If the loop is large, the two binding sites could be hundreds of kb apart. There are a few examples of such long-range interactions but they are more likely to be exceptions than rules.
In most cases the loops are more local. They form when a bound transcription factor contacts the transcription initiation complex. The idea is that binding the transcription factor anywhere in the vicinity of the promoter increases the local concentration making it likely that there will be contact between the transcription factor and the transcription complex. The classic example of a loop is the one formed by lac repressor when it contacts two separate operators (O1 and O2) [Repression of the lac Operon]. It's a model for all similar local loops. The various parameters were worked out about 25 years ago. Loop formation depends on the strength of the various protein-protein interactions and the strength of the DNA-protein interactions. The probability of forming a loop depends on the distance between the O1 and O2 binding sites, as you might imagine.
 In the case of activators and transcription complexes, the further the enhancer is away from the promoter the lower the probability that a bound transcription factor will be able to find and recognize the promoter region (in a given length of time). When the sites are far apart, it's more likely that the transcription factor will interact with other proteins that are accidentally bound in the same region. It can't be a general rule that functional sites can be 120,000 base pairs from the transcriptional start site. That's too far for serious effects in the absence of other topological constraints.
In the case of activators and transcription complexes, the further the enhancer is away from the promoter the lower the probability that a bound transcription factor will be able to find and recognize the promoter region (in a given length of time). When the sites are far apart, it's more likely that the transcription factor will interact with other proteins that are accidentally bound in the same region. It can't be a general rule that functional sites can be 120,000 base pairs from the transcriptional start site. That's too far for serious effects in the absence of other topological constraints.The authors of this paper argue that higher-order chromosome structure mediates long-range interactions but I'm not convinced that this applies to most genes.
It also can't be a general rule that the average gene is regulated by one hundred transcription factor binding sites. That doesn't make any sense. Why would most genes need this level of control? Furthermore, if mammals have evolved a higher order chromatin structure that allows for one hundred different enhancers to act at a promoter even though they are spread out over 1,000kb, then how does that work? And how do genes distinguish between genuine enhancers and spurious sites that must be littered all over that region?
Drosophila melanogaster is at least as complex as a typical mammal. Or at least it's in the same ballpark. It's genome is only 5% the size of the human genome. Why would mammals have needed to evolve so many more enhancers and so much more functional DNA in order to do something that small fruit fly genomes can do very well?
I suggested that authors should insert the following statement at the end of their papers in order to make it clear how their proposals solve the problem they are addressing.
(I/we/the authors) believe that the Deflated Ego Problem is a real scientific problem. (I/we/the authors) propose that explanation number (1/2/3/4/5/6/7) will account for the fact that we have too few genes.In this case it's #5 with a bit of a twist.
1. Exons also include 5′ and 3′ untranslated regions.
2. This value depends on a lot of assumptions and conflicting data but it's in the right ballpark.
de Laat, W. and Duboule, D. (2013) Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502:499-506. [doi: 10.1038/nature12753]

11 comments :
I dont think its a sign of an inflated ego to think mammals are more complex than flies. There are objective measures one could use such as cell type number, number of neurons or neural connectivity. If this is true then one or more of the seven possibilites list above must be the case.
One hundred enhancers per gene seems a bit much but I think its reasonable to think there are many higher levels of regulation leading to complex combinatorial control of genes/ TADs etc. I skimmed a review a few weeks ago on the tremendous complexity in TAD regulation in a single cell type and I got the impression it couldnt be explained entirely by activation/repression of loci within the TADs
I'm trying to think of observations that could confirm or refute this view. If its valid perhaps we should expect the genome to be refractory to even small perturbations. So all inversions or translocation should disrupt gene expression, even in regions that havent moved, and for no obvious reason.
The problem is "biological complexity" is not a well defined idea. Basically "complex" organisms are defined as ones most like humans and "simple" ones less like humans. Sure, you can find numbers like number of neurons that are higher in mammals than flies, but there are other numbers (such as numbers of individuals) that are higher in flies than mammals. It's entirely arbitrary which number is considered most important or "advanced".
"I dont think its a sign of an inflated ego to think mammals are more complex than flies. There are objective measures one could use such as cell type number, number of neurons or neural connectivity. If this is true then one or more of the seven possibilites list above must be the case. "
What are you really measuring by counting the number of cell types? Complexity is a concept that appeals to human beings, but what is its biological significance? Mammalians seem to have a much larger and structurally complex brain than insects, despite this fact honeybees perform complex dancing rituals using a symbolic language among a complex social hierarchy, display visual and other types of memory and are able to plan their behavior beforehand (see Zhang, S et al. 2006 for instance). "simpler" insects like Drosophila display long term memory and complex rituals and are heavily used models in behavioral genetics. Behavioral traits that require a way larger brain in mammals to be developed.
But the most interesting part of this is most of these behaviors are directed by the same pathways and transcription factors we mammals use. As a relatively simple example, the PKA-CREB axis is used by both flies and mammals to store long term memory, while a more interesting and complex one is the use of FoxP transcription factors in social communication. FoxP2 is required for language development in humans, song development in songbirds, ultrasound emission in bats and honeybee dancing(its insect homolog, FoxP).
The same goes for other traits, as we share the same basic gene circuits for heart (Tinman in flies, Nkx2-5 in humans) and eye development (Pax6) with Drosophila. And the list just goes on and on. We share a lot of our basic transcriptional building blocks and their related traits with all bilateral animals since we split in the Cambrian explosion.
There is no surprise there. We are way closer genetically to other animals than most people think, and most changes between animal clades probably lie in differential transcriptional regulation, more than brand new genetic mechanisms appeared outta nowhere.
From an evolutionary point of view, the number of cell types needed for expressing a trait or accomplishing a task is frankly irrelevant as long as the trait is adaptive. Trying to assess the complexity of behavioral and ecological interactions would probably be a more correct way imho.
I was chuckling over your list of 'serious' reasons to explain differences in complexity. Perhaps readers could offer a few more. I got one:
8. 'Higher' groups are peramorphic: Simpler animals are programmed to go through embryogenesis relatively quickly, and so have less time to develop additional levels of complexity. More complex groups like vertebrates may develop over days, but most take weeks or months. They add additional stages of development that are not identifiable in simpler forms of animals. If a nematode were genetically programmed to add additional developmental stages, taking a month to hatch from an egg, how much more complex might it be? What political party might it join?
From a YEC creationist view we are more complex because we are made in gods image. Our soul. Yet we would want our bodies to be just off the same racxk as animals in these matters.
It confirms a common blueprint from a creator for biology. Exactly what I would do if I was God(Remember Einsteins quote).
If from a creator then we should predict like atomic numbers for like needs in biology.
All biological nature is from common laws in nature with a twist at the high end.
IMO the argument that "invertebrate species studied so far tend to have more local regulatory controls, which can often be recapitulated by short transgenes" isn't cogent because the majority of published transgenic mice were produced with relatively short promoter segments. In certain cases differences of transgene expression patterns from those of the corresponding endogneous genes may be caused by missing additional regulatory sequnences.However, more often they are caused by position effect variegation. I.e. they are inserted in the vicinity of enhancers or silencers that gain influence on the regulation of the transgene. To a degree this can be omitted by flanking transgenes with insulators.Longer constructs like BAC and YAC transgenes are less prone for such effects but IMO this is likely caused by isolation from distant effects caused by sequences around the insertion site. However, in many cases the same result could be achieved by inserting the transgene into a locus (e.g. LC1) that doesn't cause positional effects.
The main problem with conventional transgenic mice is the fact that many mice derived from pro-nucleus injected oocytes don't express the transgene at all. In many cases this will be due to shortening of the transgene product before or during its insertion into the genome which employs the endogenous repair mechanisms of the cell. Thus, the fact that longer transgene constructs like BACs require less injections to obtain functional transgenes that behave like the corresponding endogenous gene may be due to the fact shortening of longer constructs is just happens further away from the sequences required for proper regulation of transgene expression.
"Yet we would want our bodies to be just off the same rack as animals in these matters."
If you're speaking as a Biblical literalist, wouldn't you have to concede that the Bible says the exact opposite, that humans were not created at the same time and as part of the same process as the animals?
A great deal of theology has been expended on the idea that mankind has a capacity for ensoulment lacking in (other) animals. If you're right, God could just have easily picked the fruit flies instead of the human race - picking would be a capricious act, not one dictated by our natures.
We were created from the same blueprint/program and our separate creation is no more relevant then bugs from birds.
the only process is a quick common design and twists to make different kinds within a single equation of biology.
Like in physics. That guy doing the same thing.
Creationism would predict and welcome a shhared atomic structure with all biology or most of it.
Ensoulment was put simply in the best body for a soul. Fruitflies can't drive or skate!
It could only be we were given the best body type as we can't have our own body type unique to us. A soul has no physical manifestation.
The world seeing such diversity and believing in evolution would imagine at the atomic level a superior/inferior score in DNA.
However no difference, because no evolution, but only common design twisting at the end biology.
Of coarse the thread is not making a creationist point. Just me.
...and so in that mega-complex organism known as the ameoba....
#9. Only humans use credit cards.
Post a Comment