
BTW, you guys did great on last week's molecule but not so good on the supplemental questions in the comments. Does everyone now know the 21st, 22nd, and 23rd amino acids?
Thus, it seems to me that there is a schisma abrew in cell evolution, with the rRNA tree and proponents of its infallibility on the one side and other forms of evidence, proponents of LGT, or proponents of a symbiotic origin of eukaryotes on the other. The former camp is well organized behind a unified view (be it right or wrong, still a view) and is arguing that we already have the answers to microbial evolution. The latter camp is not organized into castes of recognized leadership and followers, meaning that (if we are lucky) concepts and their merits, not position or power, will determine the outcome of the battle as to what ideas might or might not be worthwhile entertaining as a working hypothesis for the purpose of further scientific endeavour.The article by Norman Pace represents the side that already has the answers. He is a strong proponent of the Three Domain Hypothesis. These days, the main thrust of his argument is that we should all jump on the bandwagon or risk being left behind. I heard him speak in San Francisco last April and he sounded more like a preacher than a scientist. His article in Nature, ”Time for a Change”, is an example of the way the Three Domain Hypothesis proponents have been arguing for 20 years.
1. The gene must be universal.Only ribosomal RNA meets all three criteria, says Pace.
2. The gene must have resisted lateral gene transfer.
3. The gene must be large enough to provide useful phylogenetic information.
4. The gene must be unique, or if it isn’t, paralogues must be easily recognized.Ribosomal RNA doesn’t do so well when we add these criteria. Most bacterial genomes have multiple copies of ribosomal RNA genes. They are usually 99% similar but there are known examples of more divergent paralogues. This is not likely to be a serious problem for deep phylogeny, but it has caused problems at the species level.
5. The gene must encode a protein because it’s much more accurate to analyze amino acid sequences than nucleic acid sequences. (And easier to align.)
6. The gene must be highly conserved in order to retain significant sequence similarity at the deepest levels.
 Having declared that ribosomal RNA genes are the best choice, Pace then goes on to show us the “true”universal tree of life. As you can see, it is divided into three distinct clusters separated by long branches. The clades represent Bacteria, Archaea, and Eukaryotes; the Three Domains. The prokarotes (Bacteria and Archaea) seem to associate and the eukaryotes seem to be more distantly related.
Having declared that ribosomal RNA genes are the best choice, Pace then goes on to show us the “true”universal tree of life. As you can see, it is divided into three distinct clusters separated by long branches. The clades represent Bacteria, Archaea, and Eukaryotes; the Three Domains. The prokarotes (Bacteria and Archaea) seem to associate and the eukaryotes seem to be more distantly related. Yin, Y. and Fischer, D. (2006) On the origin of microbial ORFans: quantifying the strength of the evidence for viral lateral transfer. BMC Evolutionary Biology 2006, 6:63
[Get your free copy here]Open Access Charter
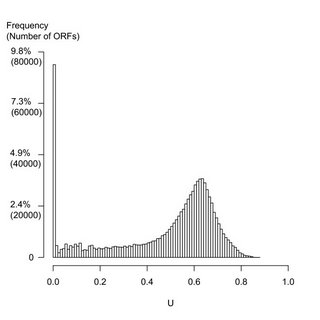 The figure shows the distribution of all 818,906 ORF's in 277 sequenced prokaryote genomes. (A typical genome has about 3000 genes.) The bottom axis represents the frequency of each of the putative genes in the database. The tall bar at the extreme left-hand side shows the number of ORF's that are only found in a single species. These are the ORFans. There are almost 80,000 of them; or, about 280 per genome. This is what the paper is all about.
The figure shows the distribution of all 818,906 ORF's in 277 sequenced prokaryote genomes. (A typical genome has about 3000 genes.) The bottom axis represents the frequency of each of the putative genes in the database. The tall bar at the extreme left-hand side shows the number of ORF's that are only found in a single species. These are the ORFans. There are almost 80,000 of them; or, about 280 per genome. This is what the paper is all about. Andrew Brown over at Helmintholog points us to Freecell fanatic about a man who has played all 32,000 games three times, and is working his way through the fourth round. You probably don't want to know which games can be won by never using the free cells, but I bet you want to know the one and only game that can't be won at all!
Andrew Brown over at Helmintholog points us to Freecell fanatic about a man who has played all 32,000 games three times, and is working his way through the fourth round. You probably don't want to know which games can be won by never using the free cells, but I bet you want to know the one and only game that can't be won at all! 
 There's been a discussion on talk.origins about calico cats—do they have to be female? The color pattern is an interesting combination of sex-linked genetics and epigenetics. Epigenetics is the inheritance of characteristics other than nuleotide sequence. In this case, it's inheritance of an inactivated X-chromosome.I used calico cats as an example in the Moran/Scrimgeour et al. textbook (1994) published by Neil Patterson/Prentice Hall. Here's an excerpt from that book.
There's been a discussion on talk.origins about calico cats—do they have to be female? The color pattern is an interesting combination of sex-linked genetics and epigenetics. Epigenetics is the inheritance of characteristics other than nuleotide sequence. In this case, it's inheritance of an inactivated X-chromosome.I used calico cats as an example in the Moran/Scrimgeour et al. textbook (1994) published by Neil Patterson/Prentice Hall. Here's an excerpt from that book.

 Tonight is hockey night in Canada. We get to watch Don Cherry on TV.
Tonight is hockey night in Canada. We get to watch Don Cherry on TV. Tonight, the Leafs play Boston at the Air Canada Centre. Some of you might recall that Boston barely squeaked out an overtime victory last time they played. Boston is home to Boston University, MIT, and some other schools.
Tonight, the Leafs play Boston at the Air Canada Centre. Some of you might recall that Boston barely squeaked out an overtime victory last time they played. Boston is home to Boston University, MIT, and some other schools.It comes down to this: no matter how many molecules you can produce with early earth conditions, plausible conditions, you're still nowhere near producing a living cell.You can't make this stuff up. And you wonder why we call them IDiots?
And here's how I know: If I take a sterile test tube and I put in it a little bit of fluid with just the right salts, just the right balance of acidity and alkalinity, just the right temperature - the perfect solution for a living cell, and I put in it one living cell, this cell is alive, it has everything it needs for life. Now I take a sterile needle and I poke that cell, and all its stuff leaks out into this test tube, you have in this nice little test tube all the molecules you need for a living cell, not just the pieces of the molecules but the molecules themselves, and you can't make a living cell out of them.
You can't put Humpty Dumpty back together again. So what makes you think that a few amino acids dissolved in the ocean are going to give you a living cell? It's totally unrealistic.