For many of us, Doty's major contribution to molecular biology was his study of DNA renaturation with his long-time post-doc and collaborator, Julius Marmur (1926 - 1996)1, a graduate of McGill University in Montréal, Canada. The paper that most of us remember is Marmur and Doty 1962: "Thermal Renaturation of Deoxyribonucleic Acids." This was the first time that the renaturation of complex DNA had been studied in detail and the results have led to many of the common techniques in use today.
THEME
Deoxyribonucleic Acid (DNA)DNA is normally double-stranded and the two strands are held together by weak interactions, chiefly stacking interactions and hydrogen bonds [DNA Denaturation and Renaturation and the Role of Hydrogen Bonds and Stacking Interactions].
The strands can be separated by heating the DNA to the "melting temperature." What Marmur and Doty showed is that the two strands could, under the appropriate conditions, come back together to form a fully base-paired double helix. This is called renaturation.
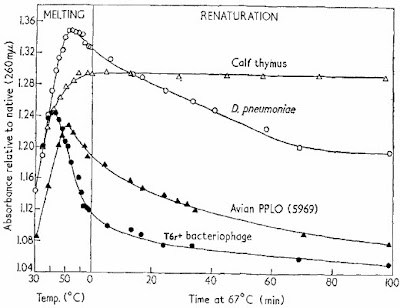
The key parameters for renaturation are time, temperature, and ionic strength and one of the key variables is the source of DNA. The first figure in the Marmur and Doty (1962) paper shows what renaturation looks like.
Single-stranded DNA absorbs much more UV light than double-stranded DNA so you can follow denaturation and renaturation by simply measuring UV light absorbance in a spectrophotometer. The figure shows that the DNA molecules "melt," or denature, as the temperature is raised. When the temperature was lowered to 67°C they could follow the formation of double-standed DNA over time. For virus and bacteriophage DNA (solid triangles and circles) most of the DNA seemed to renature in about one hour. In the case of bacterial DNA (D. pneumoniae, open circles) the process was much slower and for cow DNA (open triangles) hardly any renaturation was seen in 100 minutes.
Marmur and Doty showed that the optimum temperature for renaturation was about 25°C below the melting temperature. This was the temperature that prevented inappropriate base pairing with mismatches while still allowing true complementary strands to come together. Renaturation was more efficient in the presence of high concentrations of salt (0.3M NaCl) because the negatively charged phosphate groups had to be shielded.
Did the renaturated DNA truly represent re-formation of intact DNA molecules? Marmur and Doty proved this by showing that the pneumoniae DNA regained the ability to transform bacteria.
There are two important variables. First, the overall DNA composition in terms of GC content is important. DNA with a high GC content denatures at higher temperature and also requires a higher temperature for renaturation. We now know that this effect is due to increased stacking interactions between GC base pairs (and not the presence of three hydrogen bonds) [DNA Denaturation and Renaturation and the Role of Hydrogen Bonds and Stacking Interactions].
The second variable is DNA complexity as shown in the first figure. Cow DNA is much more complex than bacterial DNA so it takes much longer for the complementary strands to find each other in a solution of cow DNA than in a solution of bacterial DNA at the same concentration. By studying the rate of DNA renaturation as a function of time and initial concentration (Co) you can determine the genome size and how much of the DNA is single copy and multi-copy. This is the basis of Cot curves and it led to the discovery of repetitive DNA and junk DNA in eukaryotic genomes.
There are many other common techniques that rely on the pioneering work of Marmur and Doty. They include in situ hybridization, Southern blots, and the priming of PCR reactions.
On a personal note, Paul Doty is my scientific grandfather. I got my Ph.D. in 1974 with Bruce Alberts and he got his Ph.D. in 1965 with Paul Doty.
1. See the obituary in Nature by Paul Doty: Julius Marmur (1926 - 1996).
Marmur, J. and Doty, P. (1962) Thermal Renaturation of Deoxyribonucleic Acids. J. Mol. Biol. 3:585-594. [PubMed] [doi: 10.1016/S0022-2836(61)80023-5]



9 comments :
We now know that this effect is due to increased stacking interactions between GC base pairs (and not the presence of three hydrogen bonds)
Not so simple, I am afraid. Poly(dI-dC) melts at much lower temperature than poly(dG-dC). If the stacking is all that matters, there would be no difference.
I don't have any data on the stacking interactions of poly (dI-dC). Do you? I wouldn't be shocked to find out that it differs from poly (dG-dC). Would you?
There's almost a two-fold difference between CG/GC and GC/GC and CG/AT has a larger stacking interaction than GC/GC.
I never said it was simple but renaturation can't have anything to do with formation of hydrogen bonds since the single strands form many more hydrogen bonds (with water) than when they are in a double-stranded form. Renaturation involves the net loss of hydrogen bonds.
Cartoons of DNA melting (such as above) inevitably show that melting the DNA also changes the linking number. For short linear DNA molecules this makes sense, but not for long linear or circularly closed molecules. In biological systems a topoisomerase would be necessary to fully denature (or renature) DNA strands, but that's clearly not happening with these in vitro melting curves. To what extent does modeling DNA as two parallel strands vs two intertwined helical strands affect the interpretation of these melting/renaturation experiments?
I see no reason why I-I stacking would be significantly different from G-G stacking. The ring is identical. An extra amino group should affect electron distribution in the ring very little. Why would you expect it to be very different?
I certainly don't expect the simple model you present to be able to account for the fact that Tm for poly(dI-dC) is comparable to that of poly(dA-dT) and HUGELY lower than that for poly(dG-dC).
There was other important work too. A colleague of mine, Ben Hall, did his graduate work in Paul Doty's lab in the late 1950s. They established that ribosomal RNA was not coding RNA and greatly clarified the role of ribosomes. I recently asked Ben whether he had been the one who called the two subunits "18s" and "28s" (after their sedimentation rates in centrifugation). He says yes.
DK,
I'm not sure what point you're trying to make. The difference in stacking interactions accounts for the differences we see in melting temperatures of natural DNA with varying base composition. There's no other explanation that I'm aware of that explains the data. Do you know of one?
If what you say about melting (dI-dC) is correct then I expect that the measured stacking interactions will confirm it. If not, the textbooks will have to be re-written (or modified).
Do you know what salt concentration the melting studies used? I ask because poly (dG-dC) forms Z DNA at high salt. I don't know about poly (dI-dC).
DNA denaturation (strand separation) is exclusively an in vitro phenomenon. It never occurs in vivo.
The in vitro experiments are usually performed with DNA that has been sheared to sizes that are less than 1000 bp in length.
During replication, transcription, and recombination, the locally unwound region of DNA rarely exceeds 30 bp and this local unwinding is promoted by negative supercoiling. That requires topisomerases, as you correctly point out.
I was talking about poly(dI) from memory but here are two papers I found from quick googling: JMB (1964), 8:452-69; JACS (2004), 126:16387-94. It's across low/normal salt.
My point is that I am skeptical of this "water can H-bond to DNA and proteins and therefore H-bonds don't play a role in their stability" argument. I know very little about DNA but you are probably aware that the same reasoning that you provide for DNA is also frequently applied to proteins. Well, it's wrong. Fact: Synthetic proteins with backbone positions that can't form H-bonds are invariably less stable (PNAS (2006), 103:2600–2604).
Same is probably true for DNA. I just don't see how stacking alone can explain the huge difference between poly(dG-dC) and poly(dI-dC). IMO, a contribution from H-bonding has to be energetically favorable as well.
DK,
My understanding is that the base exocyclic groups - like NH2 of G - can play a very large role in stacking. According to my old copy of Principles of Nucleic Acid Structure (Saenger, 1984):
"Stacked arrangements are also dominant in crystal structures of bases. It is striking to find that the stacking patterns in these bases are rather specific, with polar substituents -NH2, =N-, =O or halogen of one base superimposed over the aromatic system of the adjacent base."
This refers to stacking of free bases, not bases within DNA, but it does indicate that the lack of NH2 in inosine could have a significant effect on stacking. That said, I didn't see anything that directly addressed stacking of poly(dI-dC) vs poly (dG-dC).
Post a Comment