Ernest Major posted a nice analysis of the paper with references to the many eplanations of the origin of ORFans. I'd like to add a bit more to his description of the "problem."
Here's the primary reference ...
Yin, Y. and Fischer, D. (2006) On the origin of microbial ORFans: quantifying the strength of the evidence for viral lateral transfer. BMC Evolutionary Biology 2006, 6:63
[Get your free copy here]Open Access Charter
ORF stands for "open reading frame" a term that refers to a stretch of codons for amino acids. It means that this ORF probably identifies a protein encoding gene. In order to be meaningful, the ORF should; (a) begin with a start codon, (b) end with a termination codon, and (c) contain a minimum number of codons (typically more than 100).
In this age of genomics and bioinformatics, there are computer programs that scan both strands of DNA to identify ORF's. These are putative genes. When the first genomes were sequenced there were a lot of putative genes that matched sequences already in the database. In other words, the computer programs identified ORF's that showed significant sequence similarity to individual genes that had already been cloned and sequenced by other labs. These genomic ORF's represented genes that were homologous to known genes.
Yin and Fischer are interested in the ORF's that aren't homologous to known genes. They concentrate on bacterial (prokaryote) genomes since the coverage is more extensive. As more and more genomes were sequenced the number of new genes represented by these non-homologous ORF's declined, as expected. Today, for every new genome that's added to the database, almost 80% of the genes have been previously identified.
The surprise is that there are so many unique ORF's in every genome. These are putative genes that have no known homologues. They are ORFans. In order to determine the number of ORFans, Yin and Fischer analyzed the complete genomes of 277 bacteria. For each and every gene they ran a search against all other genes in the database. The result was the histogram shown below.
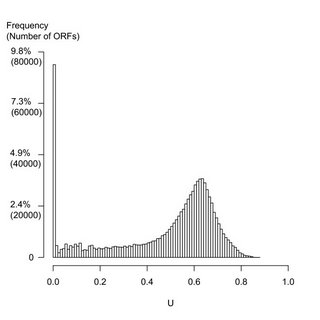 The figure shows the distribution of all 818,906 ORF's in 277 sequenced prokaryote genomes. (A typical genome has about 3000 genes.) The bottom axis represents the frequency of each of the putative genes in the database. The tall bar at the extreme left-hand side shows the number of ORF's that are only found in a single species. These are the ORFans. There are almost 80,000 of them; or, about 280 per genome. This is what the paper is all about.
The figure shows the distribution of all 818,906 ORF's in 277 sequenced prokaryote genomes. (A typical genome has about 3000 genes.) The bottom axis represents the frequency of each of the putative genes in the database. The tall bar at the extreme left-hand side shows the number of ORF's that are only found in a single species. These are the ORFans. There are almost 80,000 of them; or, about 280 per genome. This is what the paper is all about.There are some putative genes that are only present in one or two related species. These are represented by the bars at U=0.01, 0.02 etc. Some of these are also counted as ORFans since they are only present in closely related species.
As you can see, there's a broad peak of genes found in about 60% (U=0.6) of all sequenced prokaryote genomes. These represent the standard genes of metabolism. Hardly any genes are present in every single species (U=1.0). This is because the database may be incomplete, the genes may have diverged too far to be detectable, or the species is really missing that gene.
Where did the unique genes (ORFans) come from? If they are real, it seems unlikely that they sprung into existence in a single lineage. They were most likely "borrowed" from a distantly related species by a process known as lateral gene transfer. However, as more and more genomes from diverse species are added to the database it becomes worrisome that the source of these genes isn't identified.
What about viruses? It has long been known that viral genes can be incorporated into bacterial genomes so this seems like a good possibility. Yin and Fischer screened all 818,906 ORF's against the viral database to test this hypothesis. They found that only 2.8% of bacterial ORFans have detectable homologues in the viral genomes. Thus, the transfer of viral genes to bacterial genomes doesn't seem to account for all of the ORFans.
The authors discuss the problems with their experiment and urge us not to reject the viral origin hypothesis just yet. There are only 280 bacteriophage in the viral genome databse and this represents a very tiny percentage of all bacteriophage. (There may be 100 million different phage.) There are still lots of places for ORFan homologues to hide.
I think there's another problem; one that the authors are not taking seriously. It's quite possible that many of the ORFans aren't real genes at all. The computer programs that detect these ORF's are notorious for their false positives. There may be ORFan "genes" that are never transcribed or there may be ORFan "genes" that are transcribed and translated but the protein product doesn't do anything. It's an accident of evolution. In addressing this problem the authors make the common mistake of pointing to those cases where known ORFans have proven to be functional genes, while ingoring that fact most haven't. Just because some of them are real genes doesn't mean that all of them are. If most ORFans are artifacts then it's not surprising that they aren't found in other species.
2 comments :
This is a very interesting subject that seems to hardly be addressed. It would be nice if you discussed how these ORFans could be found to be functional or not. In prokaryotic organisms these ORF can be knocked out and checked for function. But what about eukaryotes?
In Drosophila, this is being done by RNA knockdown...
http://www.sciencemag.org/content/330/6011/1682.full
Post a Comment