The new completed human genome sequence contains some previously unknown large duplicatons (segmental duplications).
This is my third post on the complete telomere-to-telomere sequence of the human genome in cell line CHM13 (T2T-CHM13). There were six papers in the April 1st edition of Science. My posts on all six papers are listed at the bottom of this post.
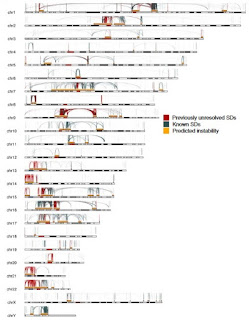
Segmental duplications (SD) consist of large regions of the human genome (>100 kb) that have been duplicated, usually by recombination errors. Some of these duplications are ancient and may be shared with closely related species but many are quite recent giving rise to polymorphisms within the species. Some of us have certain duplicated regions and others don't. There are thousands of known SDs.
The standard reference genome (CRCh38) contains a number of segmental duplications but most of us are missing some of those SDs and most of us have about 1000 extra ones that aren't in the reference genome. The assembly of the standard reference genome was complicated by the presence of SDs so it isn't clear whether it represents a typical human genome. The new T2T-CHM13 sequence is assembled from very long reads and, furthermore, the DNA is esentially haploid so it was easier to recognize SDs and other genomic rearrangements. (Most genome sequences are from diploid cells and the sister chromosomes may differ in the locations of insertions and deletions making it difficult to assemble a single complete genome that represents both copies.)
Vollger, M.R., Guitart, X., Dishuck, P.C., Mercuri, L., Harvey, W.T., Gershman, A., Diekhans, M., Sulovari, A., Munson, K.M. and Lewis, A.M. et al. (2021) Segmental duplications and their variation in a complete human genome. Science 276:55. [doi: 10.1126/science.abj6965]
Despite their importance in disease and evolution, highly identical segmental duplications (SDs) are among the last regions of the human reference genome (GRCh38) to be fully sequenced. Using a complete telomere-to-telomere human genome (T2T-CHM13), we present a comprehensive view of human SD organization. SDs account for nearly one-third of the additional sequence, increasing the genome-wide estimate from 5.4 to 7.0% [218 million base pairs (Mbp)]. An analysis of 268 human genomes shows that 91% of the previously unresolved T2T-CHM13 SD sequence (68.3 Mbp) better represents human copy number variation. Comparing long-read assemblies from human (n = 12) and nonhuman primate (n = 5) genomes, we systematically reconstruct the evolution and structural haplotype diversity of biomedically relevant and duplicated genes. This analysis reveals patterns of structural heterozygosity and evolutionary differences in SD organization between humans and other primates.
The T2T-CHM13 genome contains 208Mb of unique non-repetitive SDs (including an estimate of the Y chromosome sequence). There are significant SDs in the ribosomal RNA clusters bringing the total amount of SDs in a typical human genome to about 7% of the total sequence. The fact that these SDs are polymorphic suggests a dynamic genome with frequent duplications and deletions that, to a first approximation, don't appear to have any effect on fitness.
Note that in addition to the SDs studied in this paper, there are unique (non SD) regions of the genome that are missing in some individuals suggesting that it is junk DNA. About 7% of the unique sequences in the entire genome can be deleted without noticeable effect although no two humans differ by more than 1% in the unique regions (see Bergström et al., 2020).
Vollger et al. identified 33 new inversion polmorphisms bringing the total number to 62 known inversion polymorphisms. These are regions of the genome that have been flipped, or inverted, relative to a standard reference genome. The fact that the inversions are present in some people but not others (i.e. polymorphic) suggests that they are innocuous.
Smaller duplications can also be polymorphic and sometimes they are associated with genes. This gives rise to copy number variation and the T2T-CHM13 genome added quite a few extra examples bring the total number of known copy number variants to 1292. In terms of copy number variants, the T2T-CHM13 genome is closer to the typical genome than the standard reference genome. This is just one more bit of evidence showing that the T2T-CHM13 genome is a more faithful representation of a typical genome than CRCh38. (Most of the CRCh38 sequence is from an anonymous donor in Buffalo, New York).
The authors are clearly interested in the functions of duplicated regions and they present the data with an adaptationist bias that tends to assume functionality. There is no mention of the possibility that much of the SD and copy number variation could be unrelated to function. This approach is similar to the other papers that seem to go out of their way to avoid any mention of junk DNA.
- What do we do with two different human genome reference sequences?
- Karen Miga and the telomere-to-telomere consortium
- A complete human genome sequence (2022)
- Epigenetic markers in the last 8% of the human genome sequence
- Segmental duplications in the human genome
- Transcription activity in repeat regions of the human genome
- Genetic variation and the complete human genome sequence
Bergström, A., McCarthy, S.A., Hui, R., Almarri, M.A., Ayub, Q., Danecek, P., Chen, Y., Felkel, S., Hallast, P. and Kamm, J. (2020) Insights into human genetic variation and population history from 929 diverse genomes. Science 367:eaay5012. [doi: 10.1126/science.aay5012]

No comments:
Post a Comment