The important issue is whether alternative splicing is ubiquitous or rare. What are the evolutionary implications?
I believe that almost all of the splice variants that are routinely detected in eukaryotic cells are the product of splicing errors. (I've summarized the data on splicing errors in the Wikipedia article on Intron.) Database annotators have rejected several hundred thousand of these variants so that the typical human gene now lists only a handful of possible splice variants and very few of these have been experimentally confirmed as genuine examples of alternative splicing.
There are excellent examples of biologically relevant alternative splicing but they are confined to a small number of genes (<5%) and in almost all cases there are only a small number of alternatives (usually two) [Alternative splicing: function vs noise].
This is a controversial issue that is still being debated in the scientific literature. The important point is that respectable scientists must recognize the controversy and avoid taking dogmatic positions that are not supported by solid evidence. I'm pleased to note that more and more scientists are beginning to recognize the problem and are taking steps to present both sides of the argument. Let's look the latest review to see how this is working out.
Wright, C.J., Smith, C.W.J. Jiggins, C.D. (2022) Alternative splicing as a source of phenotypic diversity. Nature Reviews Genetics. [doi: 10.1038/s41576-022-00514-4]
A major goal of evolutionary genetics is to understand the genetic processes that give rise to phenotypic diversity in multicellular organisms. Alternative splicing generates multiple transcripts from a single gene, enriching the diversity of proteins and phenotypic traits. It is well established that alternative splicing contributes to key innovations over long evolutionary timescales, such as brain development in bilaterians. However, recent developments in long-read sequencing and the generation of high-quality genome assemblies for diverse organisms has facilitated comparisons of splicing profiles between closely related species, providing insights into how alternative splicing evolves over shorter timescales. Although most splicing variants are probably non-functional, alternative splicing is nonetheless emerging as a dynamic, evolutionarily labile process that can facilitate adaptation and contribute to species divergence.
The authors demonstrate that they are aware of the controversy over alternative splicing and they recognize that many splice variants are artifacts. However, the introduction references the standard 2008 papers (one from the Ben Blencowe lab at the University of Toronto).
Transcriptomic studies have established that AS is widespread across eukaryotes. For example, an estimated 90–95% of human genes undergo AS (Pan et al. 2008; Wang et al., 2008).
This has got to stop. There is no evidence to support such a wild claim unless you are using the term "alternative splicing" (AS) to refer to all splice variants whether they are functional or not. If that's what you mean then it would be much more accurate to say that almost all human genes produce variants due to incorrect splicing.
But let's not get too nitpicky because the authors also say ...
The extent to which AS events translate into functional protein variation is the subject of intense debate (Box 2). A number of lines of evidence suggest that the majority of observed AS events reflect splicing errors, and are neither conserved nor functional. However, there is clear evidence that a subset of AS events contribute to functional protein diversity and to regulation of protein expression levels. Moreover, the preponderance of non-functional, noisy AS events provides the potential for subsequent evolution of new function.
The Problem
Most of the paper is devoted to explaining why the production of splice variants could have evolutionary consequences. Wright et al. describe the problem in a section on "The evolution of complexity."
The genetic basis of complexity — defined as the number of distinct cell types in an organism — has been debated since comparative studies found that the total number of protein-coding genes cannot account for the increased cellular diversity observed in more complex eukaryotes. For example, both the human genome and the genome of the roundworm C. elegans have about 20,000 protein-coding genes. Numerous genomic features have been proposed to account for the poor correlation between organism complexity and total gene content (‘the G-value paradox’), including AS, microRNAs, long non-coding RNAs and non-coding DNA. Of these features, AS is a particularly attractive candidate as, by definition, it allows multiple transcripts and thus proteins to stem from a single gene.
I refer to this as the Deflated Ego Problem since it originated as a shocking revelation (to some) that the human genome didn't have more genes than "lower" animals such as nematodes.
The basic explanation has been around since publication of Stephen Jay Gould's book "Ontogeny and Phylogeny" back in 1977 and the subsequent discoveries in Evo-Devo in the 1980s. More up-to-date books include Sean B. Caroll's "Endless Forms Most Beautiful" (2005) where Sean explains it like this ...
The development of form depends upon the turning on and off of genes at different times and places in the course of development. Differences in form arise from evolutionary changes in where and when genes are used, especially those genes that affect the number, shape, or size of a structure. We will see that this has created tremendous variety in body designs and the patterning of individual structures. (p. 11)
There is no G-value paradox. We already have a sufficient explanation for why you can get tremendous diversity with the same set of genes. It may not be the complete explanation but it's sufficient to explain the differences between dozens of species of whales and their close relatives, hippopotamuses.
In fairness, the authors of this review recognize that differences in the regulation of gene expression is an important factor in generating diversity but they still think there's problem that needs a solution.
The Solution
Part of the solution involves AS (alternative splicing), which can potentially generate new protein variants.1 If AS is important, then there should be a correlation between the prevalence of AS and organism complexity.
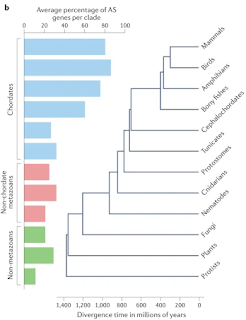
Comparative transcriptomic studies revealed extensive differences in the extent of AS between eukaryotes. Direct comparison of levels of AS across 47 diverse eukaryotic species (spanning protists, fungi, plants and animals136) revealed that the prevalence of AS (defined as the proportion of AS in multiexonic genes) was strongly correlated with organism complexity, with the highest levels in vertebrates (Fig. 1c). Importantly, this analysis accounted for differences in transcript coverage and found AS to be a strong predictor of organism complexity regardless of effective population size.
This figure does not make a lot of sense.2 If most splice variants are due to splicing errors, as the authors admit, then every gene with introns should produce splice variants in every species as long as you look hard enough.3 (The authors can't just be referring to genuine, biologically relevant, alternative splicing since there's no evidence that 80% of mammalian genes exhibit such an effect.) Besides, the latest data shows that 94% of C. elegans protein-coding genes are alternatively spliced [Alternative splicing in the nematode C. elegans] so that kind of blows that speculation out of the water!
The rest of the paper covers some examples of alternative splicing producing differences in phenotype and a few cases where closely related species show a difference in the production of splice variants. The main theme is that the evolution of differences in alternative splicing could be the underlying cause of some adaptation. The fact that production of splice variants is not conserved, as you would expect if they were splicing errors, is taken to mean that alternative splicing can evolve rapidly.
Studies indicate that splicing diverges faster than gene expression, reinforcing the view that the two processes provide alternative, complementary routes to rapid adaptation. Comparisons of organ transcriptomes from multiple vertebrate species found that AS patterns have rapidly diverged during vertebrate evolution, with splicing variation between species exceeding within-species variation across tissues.
These sentences suffer from a lack of perspective. If most of the splice variants detected in different species are due to splicing errors then comparisons between the transcriptomes of different species have to take into account the depth of coverage and whether or not the variants are due to somatic cell mutations. We're not just talking about evolution and allele frequency changes. I wish the authors, and the reviewers, had been more rigorous in analyzing the evolutionary implications of splicing errors.
The main point seems to be similar to arguments made about pervasive transcription and transposon sequences (and junk DNA in general). The idea is that the presence of these effects could make the species more likely to evolve.
It has been suggested that, by largely evolving under neutral conditions, AS can rapidly evolve and provide a route for existing genes to acquire new functions and thus adaptive benefits ....
Recent studies support the idea that AS is an important contributor to adaptive evolutionary change, interacting with other forms of genetic variation, such as transcriptional regulation. Much like mutations in the genome more broadly, the majority of splicing variation is probably neutral or mildly deleterious, representing biological noise rather than functional variation. However, this noise might represent useful standing genetic variation that could later be harnessed by selection to produce functional variants. Understanding how drift and selection interact to shape patterns of AS will be fundamental to establishing the role of alternative splicing in driving both phenotypic variation and complexity.
This argument suffers from the same teleological objections that challenge all of the other speculations about evolvability. And, like those other arguments, it suffers from a lack of evidence. While there may be a few examples of new functions that arose from splicing errors, it doesn't look like this is a common enough occurrence to warrant a grandiose speculation.
1. Alternative splicing is not confined to protein-coding genes but most papers, including this one, focus almost all of their attention on this subset of genes.
3. It reminds me of a dog-ass plot.
3. Of course, the frequency of splicing errors will depend on a number of factors that might be related to complexity, such as the number of different tissues and the size of the organism. (Bigger organisms have longer lives and more cell divisions and some of the splicing errors are due to somatic cell mutations.) Thus, it's easier to detect splicing errors in some species than in others, especially if you are looking at tissue culture cells.


I suggest that any claim regarding extensive alternative splicing has to be corroborated by Northern blotting. In most cases only one ornery few transcripts of the gene of interest will be identified.
ReplyDeleteI think this is a pretty fair review of the article. There was a lot I liked in the paper. The examples of AS functionality were interesting (I highly recommend the story of TBXT and how humans lost their tales as a - recent - example of the sort of happy accidents that can arise because of AS), there was a section on the arguments for and against the functional relevance of AS, and both the abstract and conclusions suggested that most detected AS is likely to be noise.
ReplyDeleteI was surprised with the section on the G-value paradox though. I know it used to be a mainstay of all AS articles, but I thought we had got beyond that.
Humans may have lost their tails, but I don't think that they have "lost their tales". (What a felicitous typo!)
DeleteI was thinking about making the "tale of the lost tails" joke while I was writing the post. I guess that's how it slipped in.
ReplyDelete