 "Pervasive transcription" refers to the idea that a large percentage of the DNA in mammalian genomes is transcribed. The idea became popular with the publication of the ENCODE results back in 2007 (Birney et al. 2007). Their results indicated that at least 93% of the human genome was transcribed at one time or another or in one tissue or another.
"Pervasive transcription" refers to the idea that a large percentage of the DNA in mammalian genomes is transcribed. The idea became popular with the publication of the ENCODE results back in 2007 (Birney et al. 2007). Their results indicated that at least 93% of the human genome was transcribed at one time or another or in one tissue or another. The result suggests that most of the genome consists of functional DNA. This pleases those who are opposed to the concept of junk DNA and it delights those who think that non-coding RNAs are going to radically change our concept of biochemistry and molecular biology. The result also pleased the creationists who were quick to point out that junk DNA is a myth [Junk & Jonathan: Part 6—Chapter 3, Most DNA Is Transcribed into RNA].
THEME:
Transcription
The original ENCODE paper used several different technologies to arrive at their conclusion. Different experimental protocols gave different results and there wasn't always complete overlap when it came to identifying transcribed regions of the genome. Nevertheless, the combination of results from three technologies gave the maximum value for the amount of DNA that was transcribed (93%). That's pervasive transcription.
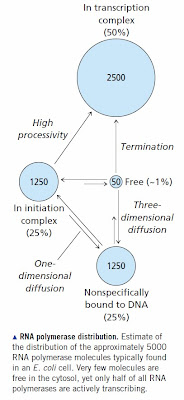
 The implication was that most of our genome is functional because it is transcribed.1 The conclusion was immediately challenged on theoretical grounds. According to our understanding of transcription, it is expected that RNA polymerase will bind accidentally at thousands of sites in the gnome and the probability of initiating the occasional transcript is significant [How RNA Polymerase Binds to DNA]. Genes make up about 30% of our genome and we expect that this fraction will be frequently transcribed. The remainder is transcribed at a very low rate that's easily detectable using modern technology. That could easily be junk RNA [How to Frame a Null Hypothesis] [How to Evaluate Genome Level Transcription Papers].
The implication was that most of our genome is functional because it is transcribed.1 The conclusion was immediately challenged on theoretical grounds. According to our understanding of transcription, it is expected that RNA polymerase will bind accidentally at thousands of sites in the gnome and the probability of initiating the occasional transcript is significant [How RNA Polymerase Binds to DNA]. Genes make up about 30% of our genome and we expect that this fraction will be frequently transcribed. The remainder is transcribed at a very low rate that's easily detectable using modern technology. That could easily be junk RNA [How to Frame a Null Hypothesis] [How to Evaluate Genome Level Transcription Papers]. There were also challenges on technical grounds; notably a widely-discussed paper by van Bakel et al, 2010) from the labs of Ben Blencowe and Tim Hughes here in Toronto. That paper claimed that some of the experiments performed by the ENCODE group were prone to false positives [see Junk RNA or Imaginary RNA?]. They concluded,
We conclude that, while there are bona fide new intergenic transcripts, their number and abundance is generally low in comparison to known genes, and the genome is not as pervasively transcribed as previously reported.The technical details of this dispute are beyond the level of this blog and, quite frankly, beyond me as well since I don't have any direct experience with these technologies. But let's not forget that aside from the dispute over the validity of the results, there is also a dispute over the interpretation.
As you might imagine, the pro-RNA, anti-junk, proponents fought back hard led by their chief, John Mattick, and Mark Gerstein (Clark et al., 2011). The focus of the counter-attack is on the validity of the results published by the Toronto group. Here's what Clark et al. (2011) conclude after their re-evaluation of the ENCODE results.
A close examination of the issues and conclusions raised by van Bakel et al. reveals the need for several corrections. First, their results are atypical and generate PR curves that are not observed with other reported tiling array data sets. Second, characterization of the transcriptomes of specific cell/tissue types using limited sampling approaches results in a limited and skewed view of the complexity of the transcriptome. Third, any estimate of the pervasiveness of transcription requires inclusion of all data sources, and less than exhaustive analyses can only provide lower bounds for transcriptional complexity. Although van Bakel et al. did not venture an estimate of the proportion of the genome expressed as primary transcripts, we agree with them that “given sufficient sequencing depth the whole genome may appear as transcripts” [2].The same issue of PLoS Biology contained a response from the Toronto group (van Bakel et al. 2011). They do not dispute the fact that much of the genome is transcribed since genes (exons + introns) make up a substantial portion and since cryptic (accidental) transcription is well-known. Instead, the Toronto group focuses on the abundance of transcripts from extra-genic regions and its significance.
There is already a wide and rapidly expanding body of literature demonstrating intricate and dynamic transcript expression patterns, evolutionary conservation of promoters, transcript sequences and splice sites, and functional roles of “dark matter” transcripts [39]. In any case, the fact that their expression can be detected by independent techniques demonstrates their existence and the reality of the pervasive transcription of the genome.
We acknowledge that the phrase quoted by Clark et al. in our Author Summary should have read “stably transcribed”, or some equivalent, rather than simply “transcribed”. But this does not change the fact that we strongly disagree with the fundamental argument put forward by Clark et al., which is that the genomic area corresponding to transcripts is more important than their relative abundance. This viewpoint makes little sense to us. Given the various sources of extraneous sequence reads, both biological and laboratory-derived (see below), it is expected that with sufficient sequencing depth the entire genome would eventually be encompassed by reads. Our statement that “the genome is not as not as pervasively transcribed as previously reported” stems from the fact that our observations relate to the relative quantity of material detected.
Of course, some rare transcripts (and/or rare transcription) are functional, and low-level transcription may also provide a pool of material for evolutionary tinkering. But given that known mechanisms—in particular, imperfections in termination (see below)—can explain the presence of low-level random (and many non-random) transcripts, we believe the burden of proof is to show that such transcripts are indeed functional, rather than to disprove their putative functionality.
 I'm with my colleagues on this one. It's not important that some part of the genome may be transcribed once every day or so. That's pretty much what you might expect from a sloppy mechanism—and let's be very clear about this, gene expression is sloppy.
I'm with my colleagues on this one. It's not important that some part of the genome may be transcribed once every day or so. That's pretty much what you might expect from a sloppy mechanism—and let's be very clear about this, gene expression is sloppy. You can't make grandiose claims about functionality based on such low levels of transcription. (Assuming the data turns out to be correct and there really is pervasive low-level transcription of the entire genome.)
This is a genuine scientific dispute waged on two levels: (1) are the experimental results correct? and (2) is the interpretation correct? I'm delighted to see these challenges to "dark matter" hyperbole and the ridiculous notion that most of our genome is functional. For the better part of a decade, Mattick and his ilk had free rein in the scientific literature [How Much Junk in the Human Genome?] [Greg Laden Gets Suckered by John Mattick].
We need to focus on re-educating the current generation of scientists so they will understand basic principles and concepts of biochemistry. The mere presence of an occasional transcript is not evidence of functionality and the papers that made that claim should never have gotten past reviewers.
1. Not just an "implication" since in many papers that conclusion is explicitly stated.
Clark, M.B., Amaral, P.P., Schlesinger, F.J., Dinger, M.E., Taft, R.J., Rinn, J.L., Ponting, C.P., Stadler, P.F., Morris, K.V., Morillon, A., Rozowsky, J.S., Gerstein, M.B., Wahlestedt, C., Hayashizaki, Y., Carninci, P., Gingeras, T.R., and Mattick, J.S. (2011) The Reality of Pervasive Transcription. PLoS Biol 9(7): e1000625. [doi: 10.1371/journal.pbio.1000625].
Birney, E., Stamatoyannopoulos, J.A. et al. (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447:799-816. [doi:10.1038/nature05874]
van Bakel, H., Nislow, C., Blencowe, B. and Hughes, T. (2010) Most "Dark Matter" Transcripts Are Associated With Known Genes. PLoS Biology 8: e1000371 [doi:10.1371/journal.pbio.1000371]
van Bakel, H., Nislow, C., Blencowe, B.J., and Hughes, T.R.. (2011) Response to "the reality of pervasive transcription". PLoS Biol 9(7): e1001102. [doi:10.1371/journal.pbio.1001102]
For what it's worth I used an Excel file in a recent paper on lincRNAs, which are fairly well defined, to add up the total genomic length of the 3300 lincRNAs they detected.
ReplyDeleteProc Natl Acad Sci U S A. 2009 Jul 14;106(28):11667-72. Epub 2009 Jul 1.
The total length was about 125 Mbp. If you double this you're still less than 10% of the genome. Of course most of this is introns that are removed from the RNAs. The lincRNA genes also overlap with protein coding genes and the 5% or so of the genome known to be conserved between multiple mammalian species. So we're still a long way from all the genome being functional.
"Genes make up about 30% of our genome"
ReplyDeleteIs it possible, that it's a typo?
In the Genetic load... and Junk in your genome: Protein encoding genes (the latter i haven't readed yet) i saw calculation, that 1.3% of our genome are genes:
2000bp/gene * 20,000genes / 3Gbp = 1.3%
Please, correct me, if i'm wrong.
Arek W. asks,
ReplyDeleteIs it possible, that it's a typo?
No. Coding regions (exons) are less than 2% of the genome but introns make up more than 25% of the genome.
A "gene" is a DNA sequence that's transcribed so this includes the introns. And it includes genes that don't encode protein.
The perplexing thing about those who believe in functionality of every detectable RNA is that they never seem to address the fact that it is not possible to not have spurious random transcription.
ReplyDeleteLarry, have you invited Mattick to join this discussion? I would be interested in knowing if he believes (as seems to be implied here) that all of the rare transcripts that can be found in high-sensitivity genome-scale studies are functional.
ReplyDeleteIf he isn't of such an opinion, then I think this argument is more of a molehill than a mountain. (Except that IDists, with their amazingly poor eyesight, are seeing Mt. Everest in all of this.)
This comment has been removed by the author.
ReplyDeleteThe reason most IDists are so committed to the idea that all the genome is functional is that for them "function" = "design" and "design" is a synonym for "individual creation of species." At least, that's how the argument looks to me. They want to be able to deny common descent (for doctrinal-Biblical reasons) as well as random mutation-natural selection and other mechanisms for evolution. So they need all the genome to be functional.
ReplyDeleteIf they want to dispute this interpretation, let someone tell us something different.
I don't have any statistics but to my best knowledge decades of analyses of inherited human diseases only lead to the discovery of mutated conventional genes. If dark matter transcripts were functional why don't they cause genetic diseases? The same is true for mutational analyses in model organisms, e.g. ENU mutagenesis in mice.
ReplyDeleteAnother question is at what frequencies dark matter transcripts occur in cells of the same tissue. According to a recent paper by Kapranov et al. (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3022773/) who congraratulate Mattick in a commnet at PLOSone (http://tinyurl.com/3dukfmm) intronic (31%) and intergenic (19%) considerably contribute to non-ribosomal/non-mitochondrial RNA in liver. However, at the same time the authors claim that this fraction contains "a large number of very long, abundantly-transcribed regions (100's of kb)". Given the fact that the exonic RNA (50% of the RNA analyzed) consists of shorter molecules (~ few kbs) this means that the frequency of RNA molecules derived from intergenic regions is at least one order of magnitude smaller than the frequency of exonic RNA. Thus, one may ask how many copies of such RNA molecules are present in a cell. If they were functional one would assume that at least one copy should be present in every individual cell of a given type. One may assume that such RNAs have a shorter half life time than exonic RNA or that that they are only expressed during a certain period of the life of a cell or only temporarily during the cell cycle. Thus, they may occur at lower frequencies than exonic RNAs. Similar questions may be asked for intronic RNA: Since introns make about 25% of the genome why is there less intronic RNA compared to exonic RNA in non-ribosomal/non-mitochondrial RNA samples? Again half-life time or cell cycle may be an issue. Still, before such questions are solved it is at least premature if not irresponsible to claim functions for dark matter transcripts.
I can't quote a ref., but I have seen data that there are mRNAs in growing yeast that are present at 1 copy per 10 cells or less.
ReplyDelete