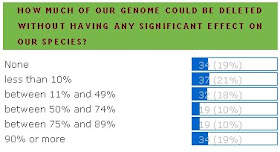
Here are the results of the most recent poll on junk DNA. Below it is the result from several years ago. I realize that the questions are different but the shift toward favoring junk DNA is still a surprise. I'd like to think it's because more and more Sandwalk readers are becoming aware of the science behind junk DNA but it could be just that more "junkers" are reading my blog.
Is there anyone out there who changed their minds over the past few years? Is The Myth of Junk DNA going to convert everyone to rejecting junk DNA? I'll have to do another poll in six months to see if the Intelligent Design Creationists are having an impact.

I'll flatly say that it was from reading this blog that I became aware that there's so much more Junk DNA that I realized. I'd thought it was a small percentage before.
ReplyDeleteI don't like the term "junk". It biases our conception of genomic clutter (things that don't have an immediate purpose), whatever the cause or reason. The idea that evolution will get rid of components it doesn't "use" is likewise misguided (at least to me). Upon reading the literature on transposons (not for the faint of heart), I'm beginning to think that higher-level regulation in the genome might depend on this "junk" to inhibit gene products, and then inhibit the inhibitors. This model is sort of like what happens in some brain circuits. If an evolutionary arms race occurs among the inhibitors (or other regulators), you might get massive amounts of genomic material that act recursively -- not really amenable to current analyses.
ReplyDeletee.racer says,
ReplyDeleteI don't like the term "junk".
You don't like the term "junk" because you don't think it's junk. Right?
I don't like the way some people misunderstand and/or abuse the term "junk DNA". I don't find anything wrong with the term itself.
ReplyDeleteI think the questions are too different to compare. I think most of the genome is non-functional in the sense that sequence does not matter; it just rides along with the bits that do, and complete replacement of any such sequence by any other similarly non-functional sequence would have no effect.
ReplyDeleteBut if we could engineer a species-wide excision of these chunks (otherwise damage to fertility due to meiotic incompatibility would be "an effect"), that still could have a significant effect on development, simply by changing the timing of the cell cycle.
Even 'true' junk, having neither an informational nor a structural role, can't just be chucked away wholesale.
@Allan Miller,
ReplyDeleteIt's a known fact that some DNA is essential even though the sequence isn't conserved. It's also a fact that the only experiment where large sections of junk DNA were deleted showed no effect (i.e. they really were junk).
Your speculation that a large percentage of "junk" DNA is essential is not based on anything we currently know about biochemistry or molecular biology. It sounds an awful lot like the sort of "special pleading" that's a characteristic of someone who has other reasons for not wanting our genome to be full of junk.
What are those other reasons? Is it because you are an adaptationist who just doesn't like the idea of non-adaptive evolution?
What are those other reasons? Is it because you are an adaptationist who just doesn't like the idea of non-adaptive evolution?
ReplyDeleteLarry,
Absolutely not, no. I perceive something of a knee-jerk reaction, there! I'm one of the ">95%" brigade. I am perfectly happy with the idea of non-adaptive evolution, and of a genome that is largely a waste of space.
I see junk accumulating by ratchet. Within the parameters of selective advantage, there is insufficient penalty for a lineage that acquires a chunk of unnecessary DNA, and insufficient reward for one that manages to shed one. The ratchet is biased in an upward direction because shedding is more likely to do damage than simply living with it.
But adaptive evolution may well be lurking somewhere, not to build junk as an adaptation, but because its presence has affected other adaptive processes. It's no different from constructive neutral evolution in that sense. What I am suggesting is that 'higher organism' developmental cycles were tuned in the light of eukaryotic c-values. So to simply flip to another, radically different c-value is not necessarily going to have a null effect. I agree that experiments done to date don't indicate such an effect. But I am considerably more cautious in my assessment of the amount of genome that can be deleted without effect (<30%) than I am in my assessment of how much is junk (>96%).
But I am considerably more cautious in my assessment of the amount of genome that can be deleted without effect (<30%) than I am in my assessment of how much is junk (>96%).
ReplyDeleteI agree with this. I think the questions are quite different, and difficult to compare.
I'm confident that >90% of the human genome is sequence-irrelevant - random substitution throughout that 90% would have no effect on survival or reproduction (ignoring potential complications during meiotic recombination).
However, the experiments involving deletions have not been deletions of more than half the genome. I don't know where the point is that massive deletion causes fitness effects, but I suspect is is somewhere around 1/3 to 2/3 of the genome, without deletion of coding sequences or other non-junk parts.
C-value and cell volume are related, and cell volume has clear effects on cell division, adult body size of multicellular organisms, tissue repair and regeneration, and other aspects of organisms with major fitness consequences. My thinking is that a change of a few percent in cell volume isn't going to do much, but a change of 50% or 80% would have large effects.
I freely admit that the above musings are based on nothing more than supposition and speculation - are there any studies out there (say, in a unicellular eukaryote, maybe yeast) in which hundreds of large deletions were inflicted upon a genome?
Although much of DNA is probably junk, having no coding or regulatory function, maybe even 90%, eliminating all of that DNA would likely have an effect on the organism, because the rates and procedures in the cell of repairing, replicating and moving about DNA are correlated with the genome's size (regardless of its content).
ReplyDeleteIf you wanted to prove a point about 90% being junk, grow numerous batches of cloned cells, each of which having a significant, but much smaller than 90%, proportion of its thought-junk DNA removed. If some batches of clones thrive which, collectively, have DNA removed which covers 90% of the original genome (or even just a large proportion of it), then that's a strong suggestion in favour of much DNA being junk.
I am curious about the statements that mass deletions "must have" an effect. What are we basing this assumption on? Going the other way and substantially increasing the amount of DNA (polyploidy in fish and amphibians) does not seem to have any sort of mass effect on the species pairs...
ReplyDeleteTheBrummell says,
ReplyDeleteC-value and cell volume are related, and cell volume has clear effects on cell division, adult body size of multicellular organisms, tissue repair and regeneration, and other aspects of organisms with major fitness consequences. My thinking is that a change of a few percent in cell volume isn't going to do much, but a change of 50% or 80% would have large effects.
I'm sure you're aware of The Onion Test. How does it apply to your thinking about this subject?
I'm sure you're aware of the fact that there are species of frog that differ by eight-fold in the amount of DNA in their genomes (2C, 4C and 8C). I don't think you'd be able to tell the difference if you examined the morphology and behavior of those frog species. How dies that fact fit in with your speculation?
Good points, Dr. Moran. I'm not particularly familiar with the multiploid frogs, but I would have to look up what data there are regarding development rates, growth rates, size-and-age at metamorphosis, et cetera, to speculate further on the relationships between cellular DNA contents, cell volume, and life-history. Thanks for that, I'll have to ponder this some more.
ReplyDeleteRegarding the Onion Test, I am indeed (under the impression that I am) familiar with it... it's been a few years since I last thought about it in any serious way. If I read it correctly, it is primarily an argument against assigning function to large amounts of putatively-and-probably-junk DNA when some function is discovered for some small piece; it's a caution about unreasonable extrapolation. The question about why can some organisms survive with much more or much less DNA per cell than other organisms is quite useful in this context. But it does not negate the relationship between cellular DNA contents and cell volume, and the life-history trait variation associated with cell volume variation.
I am still thinking about junk DNA in two distinct ways - as DNA that could be duplicated or deleted without significant effects on an organism's fitness, and as DNA that could not be so doubled or removed but could have its sequence completely substituted at random without fitness consequences.
Would you say a piece of DNA is junk if deletion causes mortality or reproductive failure, but random substitution does not?
@TheBrummell,
ReplyDeleteHere's the link to the Onion Test.
It asks you to explain why three closely related species of onion have genomes of 7 pg, 17 pg, and 31.5 pg.
Larry said:
ReplyDeleteI don't think you'd be able to tell the difference if you examined the morphology and behavior of those frog species.
I am not familiar with the frog example either, but I know that polyploidy does have an effect in several plant species. Autopolyploids have an overall increase in cell size, increased thickness of leaves, increased retention of water, slower growth, delayed flowering and flowering over a longer season. So, like Allan Miller and TheBrummell, I suspect that there will be a phenotypic effect of deletion of massive amounts of non-coding DNA.