 "What do protein crystallographers dream of?" is the question asked by Ananyo Bhattacharya in an article published in Nature [Protein structures: Structures of desire].
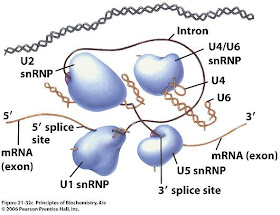
"What do protein crystallographers dream of?" is the question asked by Ananyo Bhattacharya in an article published in Nature [Protein structures: Structures of desire]. The structures of many protein complexes have been determined but crystallographers have a list of holy grails that, so far, have eluded them. It's an interesting list and one that I mostly agree with. Can you identify the structure shown here in cartoon form?
One glaring omission is pyruvate dehydrogenase. Lot's of people want to see that structure. Other notable omissions include complex I of the membrane-associated electron transport chain and the protein import complex of the endoplasmic reticulum. Don't protein crystallographers dream of those?
Isn't it the spliceosome complex in action? I do wonder, however if the process of crystallography is too inefficient for the studies we will require in future. Its had its stunning successes but it doesnt easily lend itself to the sort of factory line analysis that is currently required.
ReplyDeleteMore GPCRs...
ReplyDeleteLarry,
ReplyDeletepiruvate carboxylase, complete with all four domains, has been solved:
http://www.ncbi.nlm.nih.gov/pubmed/17717183
(PDB ID 2QF7). There are still few things to figure out but the basic mechanism is all clear!
Another glaring omission is a structure of actomyosin. Just two proteins are the basis for perhaps the most important kind of motility we know. Structures of both proteins individually are well known and an abundance of models and low resolution data is available - yet we still have basically no molecular view of 1) how actin interacts with actin to produce filaments, 2) how myosin interacts with actin and how this interaction is converted into mechanical force.
Unlike the megastructures listed, the problem is intrinsic - actomyosin forms a gel "by design", something that is totally incompable with the very idea of crystallization.
Back to megastuctures, centrioles *seem* to be symmetric and well-ordered enough but we still have no ability to obtain them in the form amenable to crystallization. Crystallographers are working on it, though :-)
Re: GPCRs. Nah... The holy grail was found with rhodopsin, the rest (that is surely slowly coming), is going to be incremental.
Oops, I am afraid my comment about pyruvate carboxylase will look like non sequitur. I meant to add that the basic principles observed for PC are likely to be similar to for PDH - intermolecular transfer and allosteric regulation.
ReplyDeleteThis comment has been removed by the author.
ReplyDeleteNot for drug discoverers. Even though rhodopsin was a landmark, you need to work in drug discovery to appreciate how much we will appreciate more GPCR structures. Most GPCRs of interest in drug discovery can have a sequence similarity of 15-20% with rhodopsin. With this kind of similarity, homology modeling can be and is a nightmare and lacks predictive power.
ReplyDeleteAnd as you may know, currently the rave is not really the rhodopsin structure but the beta2-adrenergic structure.
Mostly I dream about members of the opposite sex, but that's just me.
ReplyDelete"The crystals look beautiful under the microscope."
ReplyDeleteThat's primarily due to optical properties of polystyrene under polarized light.