The figure below is taken from Doolittle's Scientific American article "Uprooting the Tree of Life" (February 2000). © Scientific American

Doolittle has a new paper out in PNAS (Doolittle and Bapteste, 2007). In that paper he restates his ideas about the web of life and emphasizes the fact that most of us are making unsubstantiated assumptions about the treelike structure of life. This is not a criticism of evolution—far from it—but as you might expect it has attracted the attention of anti-science writers such as Casey Luskin.
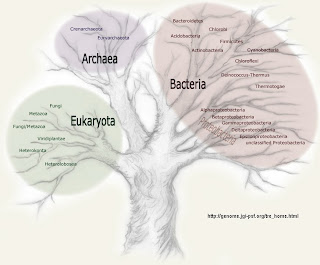 Doolittle and Bapteste (2007) are opposed to the bifurcating tree of life such as the one shown on the Dept. of Energy (USA) Joint Genome Initiative website [JGI Microbial Genomes]. They say,
Doolittle and Bapteste (2007) are opposed to the bifurcating tree of life such as the one shown on the Dept. of Energy (USA) Joint Genome Initiative website [JGI Microbial Genomes]. They say,The meaning, role in biology, and support in evidence of the universal "Tree of Life" (TOL) are currently in dispute. Some evolutionists believe (i) that a single rooted and dichotomously branching representation of the relationships between all life forms is appropriate (at all levels above species), because it best represents their history; (ii) that we can with available data and methods reconstruct this tree quite accurately; and (iii) that we have in fact done so, at least for the major groups of organisms. Other evolutionists question the second and third of these beliefs, holding that data are as yet insufficiently numerous and phylogenetic models as yet insufficiently accurate to allow reconstruction of life's earliest divisions, although they do not doubt that some rooted and dichotomously branching tree can in principle represent the history of all life. Still other evolutionists, ourselves included, question even this most fundamental belief, that there is a single true tree. All sides express confidence in their positions, and the debate often seems to be at an impasse.The argument is long and complex but the essence is that we need to abandon our assumption that the tree of life can be represented by: (i) a unique hierarchical pattern, (ii) the historical record can be best represented by a branching pattern, and (iii) natural selection is the primary cause of speciation.
We've pretty much abandoned the third point ...
As to this third possibility, modern evolutionists accept the uncoupling of selection from divergence, not only at the molecular level (the neutral theory) but in certain models for speciation, without seeing the Darwinian (or at least the neo-Darwinian) theory as refuted (21, 22). We have come to appreciate the plurality of evolutionary processes of lineage diversification. But most of us hold on to the first two tenets, that there is a real and universal natural hierarchy, and that descent with modification explains it, in much the same way as Darwin did. We may be process pluralists, but we remain pattern monists.This brings us to the title of their paper Pattern Pluralism and the Tree of Life Hypothesis. Doolittle and Bapsiste want us to not only be pluralists with respect to the mechanism of evolution but also with respect to the pattern of evolution.
Doolittle maintains that lateral gene transfer (LGT) is so common that it's impossible to construct a reliable bifurcating tree to represent the actual history of life. In other words, a Tree of Life is not only technically difficult but impossible in theory as well. This problem extends to all branches of the prokaryotic tree including the major divisions. Even the existence of two prokaryotic domains is questionable. Rooting the tree of life is out of the question.
I'm a big fan of Ford Doolittle—after all, he's an honorary Canadian! I certainly agree with him about the demise of the Three Domain Hypothesis. (Most people seem to have missed the death announcement.) I also agree with him that early evolution is more like a web of life than a tree of life. Nevertheless, I think he goes too far. Lateral gene transfer is an important, and common, phenomenon but I don't think it's quite as prevalent as he makes out. I still think that a bifurcating tree can be used to represent most species evolution after about 2.5 billion years ago.
Doolittle, W.F. and Bapteste, E. (2007) Pattern pluralism and the Tree of Life hypothesis. Proc. Natl. Acad. Sci. (USA) 104:2043-2049. [PubMed]
I agree with you, Larry. I also believe that lateral transfer is significant but still rare enough that an overall tree of life structure is still maintained.
ReplyDeleteHowever, I also think that in early evolution, lateral trasfer was more common and, thus, makes the "trunk" of the tree look more like a fuzzy swamp.
Larry,
ReplyDeleteYou might be interested in the following paper just out in PNAS (its also open access so everyone can read):
Global extent of horizontal gene transfer
http://www.pnas.org/cgi/content/full/104/11/4489
I wonder why Luskin left this bit of the paper out:
ReplyDelete"And it should not be an essential element in our struggle against those who doubt the validity of evolutionary theory, who can take comfort from this challenge to the TOL only by a willful misunderstanding of its import."
Adding to what Larry wrote, here's a nice example of how the increasing availability of whole-genome sequences is being used to correct the oversimplified pictures derived from comparing single genes like 16S rRNA: http://tinyurl.com/yoq5u2
ReplyDeleteSee the last paragraph of the discussion for how the data undermine the three domains.
Larry is obviously infinitely more up to speed on this literature than I am so his comments on this Konstantinidis and Tiedje paper would be interesting.
Does the extent of gene transfer really suggest there aren't three domains of life? Archaea do differ in very significant ways from bacteria and eukaryotes; bacteria differ from archaea and eukaryotes in significant ways; eukaryotes differ from archaea and bacteria in significant ways. There are similarities yes from a multitude of gene transfers between all domains making a trunk impossible to see but that doesn't mean archaea should be grouped with bacteria.
ReplyDeleteponderingfool says,
ReplyDeleteArchaea do differ in very significant ways from bacteria and eukaryotes; bacteria differ from archaea and eukaryotes in significant ways; eukaryotes differ from archaea and bacteria in significant ways.
Bacteria are a large and diverse group. Cyanobacteria differ in many ways from myxobacteria and myocoplamas are very different from their relatives, the soil bacteria, endospora.
Within the group you refer to as bacteria is the clade known as anoxic thermophiles (Thermogotae) which is barely distinguishable in many ways from archaebacteria.
The point is that there are many different kinds of bacteria and the differences between archaebacteria and most other groups is no greater that the differences between the groups that aren't called archabacteria.
The differences between prokaryotes and eukaryotes are more substantial and more stable. That's why there's a movement to rethink early evolution with a view to putting the split between eukaryotes and prokayotes deep in the tree at, perhaps, 3 billion years ago.
The point is that there are many different kinds of bacteria and the differences between archaebacteria and most other groups is no greater that the differences between the groups that aren't called archabacteria.
ReplyDeleteThe differences between prokaryotes and eukaryotes are more substantial and more stable. That's why there's a movement to rethink early evolution with a view to putting the split between eukaryotes and prokayotes deep in the tree at, perhaps, 3 billion years ago.
****************************************
The differences between archaea and bacteria though are pretty substantial. Different membranes. Different means of making Gln-tRNA(Gln). Archaea use Met-tRNA and not fMet-tRNA to initiate translation like all known archaea. I would expect large differences within archaea and bacteria given the selective pressures they are under. The ribosomes are also different and RNA polymerase and transcription factors. Those are some major differences that can't just be brushed aside. Similarities between Thermotoga and archaea come down to adaptations to survive at higher temperatures, exactly the type of genes we would expect to transfer.
Bacteria and archaea exist in all types of environments. Given this and the low barriers to gene transfer, I would expect based on whole genome comparisons some bacteria looking more archaeal like and vice versa.
Given the small sampling of the biodiversity especially of bacteria and archaea , I don't expect our current theories to stand but given all the evidence going back to the prokaryotes/eukaryote divide is going to suffice.
The ssu rRNA tree on which the original 3-D system was based presumably reflects a phylogeny of ribosomes (three clearly separate lineages, separate for a very long time)--the question is whether it accurately reflects the phylogeny of the cells (and/or genomes) that contained those ribosomes (ribosomal genes), yes?
ReplyDeleteGiven the number of different gene products that have to interact to pull off successful translation, I'd expect ribosomal genes would be far less likely to be exchanged laterally between dissimilar cells than just about any other type of gene--enzymes, certainly should be more readily shared, and HSPs too (as they function by interacting with various & sundry proteins anyway...I think).
I covered the "complexity" rationalizations in two separate posts The Three Domain Hypothesis (Part 4) and The Three Domain Hyptothesis (part 5).
ReplyDeleteIn the second article I discussed the three reasons why the complexity argument is wrong.
1. The facts don't support it. There are many translation proteins that don't show three domains.
2. The logic is flawed. Trees are based on the parts of a sequence that change and those, by definition, are not the conserved regions that interact.
3. There's no reason to assume that genes for groups of interacting proteins have to identify a "core" that represents the cell phylogeny. One could just as easily argue that the evidence indicates that these genes are always transferred together.
The sad thing about this debate is that is has been characterized by a notable lack of rationality. Most of the arguments supporting the Three Domain Hypothesis were based more on wishful thinking and subjective examination of selected facts than on real scientific reasoning.
Larry which aminoacyl-tRNA synthetases are you talking about? There are two LysRS enzymes (one class I and the other class II). Some organisms have both along with PylRS. GlnRS is not commonly found in bacteria and is absent in all known archaea. GlnRS appears to have evolved from a eukaryotic GluRS. A few bacteria such as E. coli have acquired this eukaryotic tRNA synthetase. GluRS follows a three domain phylogeny, so does AspRS. AsnRS appears to have evolved from eukaryotic/archaeal type of AspRS (as does AsnA).
ReplyDeleteYou also have the ncRNAs. Only the SRP, RNaseP, tRNA and SRP are found in archaea, batteria and eukaryotes. snoRNAs are found in archaea and eukaryotes. Then there are ncRNAs only found in certain domains. rRNA, RNaseP, and SRP support a three domain phylogeny.
And the arguements you are making do not maintain the grouping of bacteria with archaea. You are arguing that it can't be known in which case the old grouping is not supported either.
Thanks for linking those earlier posts. Very interesting stuff, and I appreciate your summarizing all that current thinking.
ReplyDeleteIf the deep phylogeny of cells/organisms/genomes is, then, unknowable, what CAN we say?
Eukarya seems unquestionably monophyletic (leaving mitochondria and plastids aside, of course) and so we are left with a huge, diverse, possibly paraphyletic (but we'll never know) and possibly even polyphyletic??? (but we'll never know) Domain Monera?
This is rather unsatisfying, but if that is the situation, so be it.
PonderingFool says,
ReplyDeleteAnd the arguements you are making do not maintain the grouping of bacteria with archaea. You are arguing that it can't be known in which case the old grouping is not supported either.
There are many genes that support clustering of archaebacteria within other bacteria. There are some that support a distant relationship between archaebacteria and other bacteria. There are very few that support a clustering of archaebacteria with eukaryotes—this include the ribosomal RNA phylogenies.
The results demonstrate that no consensus is possible and that's why the Three Domain Hypothesis has been abandoned in favor of a web of life.
The supporters of the Three Domain Hypothesis seem reluctant to accept the totality of the scientific evidence so they tend to focus on a few genes that support their favorite hypothesis and reject or ignore those that don't. They put forward arguments attempting to prove that their genes are somehow better than others at identifying the true tree of life.
So far, those arguments have not stood up to close scrutiny. The only reasonable conclusion is that there is no universal tree of life. Why are you so reluctant to accept this?
Larry I am fine with a web and not a single trunk. My problem is keeping the old prokaryotic grouping of bacteria and archaea in terms of phylogenetics. Prokaryotes is fine defining as not eukaryotic but the the archaea are different enough that I would say they are separate from bacteria. Where eukaryotes come is hard to say because they seem to e a fusion of some sort between bacteria, archaea and maybe proto-euk.
ReplyDeleteAnd in your death of the three domain hypothesis, what aaRSs were you talking about that did not fit? Curious which ones.
ponderingfool says,
ReplyDeleteWhere eukaryotes come is hard to say because they seem to e a fusion of some sort between bacteria, archaea and maybe proto-euk.
When you make a statement like this you are assuming that archaebacteria and other bacteria are in different domains. Once you let go of that idea it becomes easier to understand the origin of eukaryotes. The problem in the recent past has been to resolve the apparent "paradox" that resulted when the majority of eukaryotic genes were found to be closer to other bacteria than to archaebacteria. This was contrary to the prediction of the Three Domain Hypothesis.
Instead of interpreting this as refuting the Three Domain Hypothesis, a number of "fusion" senarios where invented. Most of them aren't valid once you realize that the root of the tree does not lie between archaebacteria and other bacteria.
And in your death of the three domain hypothesis, what aaRSs were you talking about that did not fit? Curious which ones.
Turns out that none of the aminoacyl-tRNA synthase genes are good phylogenetic markers. According to Ludwig and Schleifer (2005) (see The Three Domain Hypothesis (part 4)) there are only four of them (aspartyl-, leucyl-, tryptophanyl-, and tryrosyl-) that support the hypothesis.
Of the remaining tRNA synthases, arginyl-, cysteinyl-, lysyl-, and threonyl- clearly refute the hypothesis. The others are ambiguous or show weaker rejection of the Three Domain Hypothesis.
Many of the ribosomal protein genes do not support the hypothesis and the dnaK/HSP70 genes—one of the best phylogenetic markers known—clearly and unambiguously refutes the Three Domain Hypothesis. (HSP70 is the chaperone that interacts with ribosomes during protein synthesis.)
What other genes follow a similar phylogenetic pattern as hsp70?
ReplyDeleteGiven the signficant amount of gene transfer, wouldn't is be beneficial for organisms to acquire chaperones in order to properly fold the acquired enzymes?